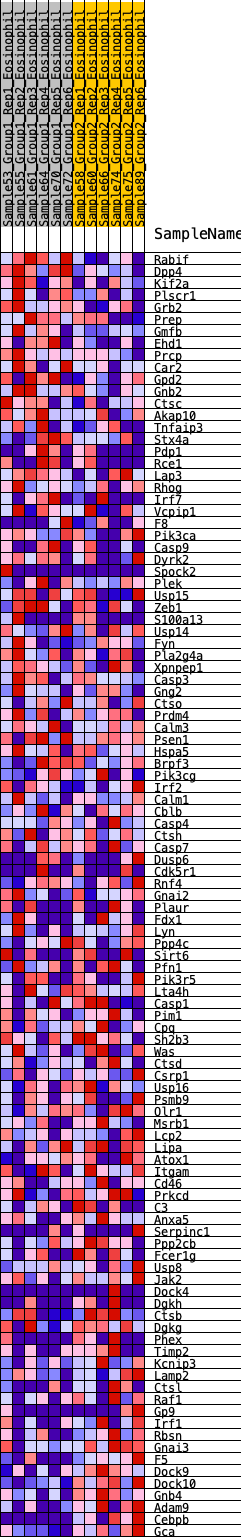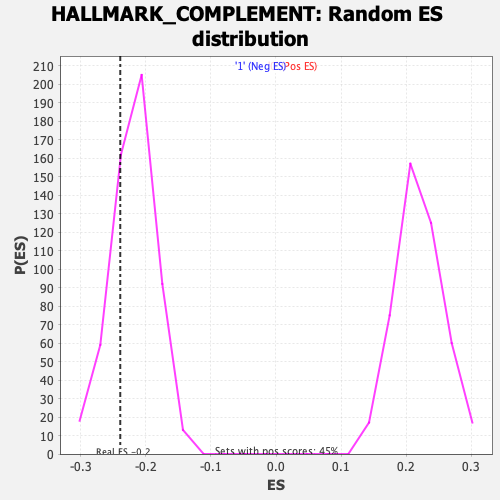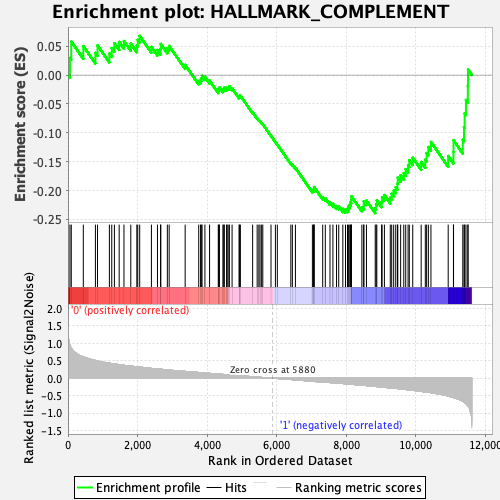
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.2392529 |
| Normalized Enrichment Score (NES) | -1.0948802 |
| Nominal p-value | 0.24043716 |
| FDR q-value | 0.501334 |
| FWER p-Value | 0.989 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rabif | 56 | 0.956 | 0.0299 | No |
| 2 | Dpp4 | 94 | 0.863 | 0.0581 | No |
| 3 | Kif2a | 440 | 0.610 | 0.0503 | No |
| 4 | Plscr1 | 788 | 0.508 | 0.0386 | No |
| 5 | Grb2 | 849 | 0.496 | 0.0515 | No |
| 6 | Prep | 1190 | 0.429 | 0.0375 | No |
| 7 | Gmfb | 1258 | 0.419 | 0.0469 | No |
| 8 | Ehd1 | 1334 | 0.409 | 0.0553 | No |
| 9 | Prcp | 1471 | 0.387 | 0.0575 | No |
| 10 | Car2 | 1609 | 0.368 | 0.0590 | No |
| 11 | Gpd2 | 1805 | 0.345 | 0.0546 | No |
| 12 | Gnb2 | 1979 | 0.327 | 0.0515 | No |
| 13 | Ctsc | 2005 | 0.325 | 0.0612 | No |
| 14 | Akap10 | 2061 | 0.318 | 0.0680 | No |
| 15 | Tnfaip3 | 2398 | 0.280 | 0.0489 | No |
| 16 | Stx4a | 2572 | 0.266 | 0.0436 | No |
| 17 | Pdp1 | 2661 | 0.257 | 0.0453 | No |
| 18 | Rce1 | 2668 | 0.256 | 0.0540 | No |
| 19 | Lap3 | 2856 | 0.238 | 0.0465 | No |
| 20 | Rhog | 2908 | 0.234 | 0.0505 | No |
| 21 | Irf7 | 3368 | 0.194 | 0.0177 | No |
| 22 | Vcpip1 | 3758 | 0.163 | -0.0102 | No |
| 23 | F8 | 3811 | 0.157 | -0.0090 | No |
| 24 | Pik3ca | 3832 | 0.156 | -0.0051 | No |
| 25 | Casp9 | 3854 | 0.154 | -0.0013 | No |
| 26 | Dyrk2 | 3936 | 0.147 | -0.0030 | No |
| 27 | Spock2 | 4070 | 0.138 | -0.0096 | No |
| 28 | Plek | 4320 | 0.118 | -0.0269 | No |
| 29 | Usp15 | 4330 | 0.117 | -0.0235 | No |
| 30 | Zeb1 | 4354 | 0.115 | -0.0213 | No |
| 31 | S100a13 | 4453 | 0.108 | -0.0258 | No |
| 32 | Usp14 | 4477 | 0.107 | -0.0240 | No |
| 33 | Fyn | 4494 | 0.106 | -0.0215 | No |
| 34 | Pla2g4a | 4559 | 0.101 | -0.0234 | No |
| 35 | Xpnpep1 | 4578 | 0.100 | -0.0213 | No |
| 36 | Casp3 | 4625 | 0.096 | -0.0218 | No |
| 37 | Gng2 | 4638 | 0.095 | -0.0194 | No |
| 38 | Ctso | 4717 | 0.089 | -0.0230 | No |
| 39 | Prdm4 | 4919 | 0.072 | -0.0378 | No |
| 40 | Calm3 | 4927 | 0.072 | -0.0358 | No |
| 41 | Psen1 | 4956 | 0.069 | -0.0357 | No |
| 42 | Hspa5 | 5308 | 0.043 | -0.0647 | No |
| 43 | Brpf3 | 5438 | 0.032 | -0.0747 | No |
| 44 | Pik3cg | 5487 | 0.029 | -0.0779 | No |
| 45 | Irf2 | 5521 | 0.025 | -0.0798 | No |
| 46 | Calm1 | 5567 | 0.022 | -0.0830 | No |
| 47 | Cblb | 5600 | 0.019 | -0.0850 | No |
| 48 | Casp4 | 5836 | 0.003 | -0.1053 | No |
| 49 | Ctsh | 5962 | -0.004 | -0.1161 | No |
| 50 | Casp7 | 6018 | -0.009 | -0.1205 | No |
| 51 | Dusp6 | 6406 | -0.037 | -0.1528 | No |
| 52 | Cdk5r1 | 6450 | -0.040 | -0.1551 | No |
| 53 | Rnf4 | 6538 | -0.048 | -0.1609 | No |
| 54 | Gnai2 | 7026 | -0.088 | -0.2000 | No |
| 55 | Plaur | 7057 | -0.091 | -0.1993 | No |
| 56 | Fdx1 | 7071 | -0.093 | -0.1971 | No |
| 57 | Lyn | 7078 | -0.094 | -0.1942 | No |
| 58 | Ppp4c | 7322 | -0.109 | -0.2113 | No |
| 59 | Sirt6 | 7397 | -0.116 | -0.2135 | No |
| 60 | Pfn1 | 7532 | -0.125 | -0.2206 | No |
| 61 | Pik3r5 | 7616 | -0.131 | -0.2231 | No |
| 62 | Lta4h | 7720 | -0.138 | -0.2270 | No |
| 63 | Casp1 | 7783 | -0.143 | -0.2272 | No |
| 64 | Pim1 | 7901 | -0.152 | -0.2318 | No |
| 65 | Cpq | 7978 | -0.159 | -0.2326 | Yes |
| 66 | Sh2b3 | 8035 | -0.163 | -0.2316 | Yes |
| 67 | Was | 8061 | -0.166 | -0.2277 | Yes |
| 68 | Ctsd | 8100 | -0.168 | -0.2249 | Yes |
| 69 | Csrp1 | 8121 | -0.171 | -0.2204 | Yes |
| 70 | Usp16 | 8145 | -0.173 | -0.2161 | Yes |
| 71 | Psmb9 | 8148 | -0.173 | -0.2100 | Yes |
| 72 | Olr1 | 8449 | -0.198 | -0.2289 | Yes |
| 73 | Msrb1 | 8503 | -0.205 | -0.2261 | Yes |
| 74 | Lcp2 | 8504 | -0.205 | -0.2186 | Yes |
| 75 | Lipa | 8580 | -0.211 | -0.2174 | Yes |
| 76 | Atox1 | 8832 | -0.235 | -0.2307 | Yes |
| 77 | Itgam | 8858 | -0.238 | -0.2242 | Yes |
| 78 | Cd46 | 8878 | -0.240 | -0.2171 | Yes |
| 79 | Prkcd | 9015 | -0.252 | -0.2198 | Yes |
| 80 | C3 | 9029 | -0.253 | -0.2117 | Yes |
| 81 | Anxa5 | 9096 | -0.261 | -0.2079 | Yes |
| 82 | Serpinc1 | 9265 | -0.278 | -0.2124 | Yes |
| 83 | Ppp2cb | 9306 | -0.283 | -0.2056 | Yes |
| 84 | Fcer1g | 9363 | -0.287 | -0.2000 | Yes |
| 85 | Usp8 | 9422 | -0.292 | -0.1945 | Yes |
| 86 | Jak2 | 9471 | -0.298 | -0.1878 | Yes |
| 87 | Dock4 | 9481 | -0.299 | -0.1777 | Yes |
| 88 | Dgkh | 9563 | -0.307 | -0.1736 | Yes |
| 89 | Ctsb | 9653 | -0.316 | -0.1698 | Yes |
| 90 | Dgkg | 9709 | -0.322 | -0.1629 | Yes |
| 91 | Phex | 9777 | -0.332 | -0.1566 | Yes |
| 92 | Timp2 | 9812 | -0.337 | -0.1474 | Yes |
| 93 | Kcnip3 | 9911 | -0.347 | -0.1432 | Yes |
| 94 | Lamp2 | 10153 | -0.377 | -0.1505 | Yes |
| 95 | Ctsl | 10272 | -0.394 | -0.1464 | Yes |
| 96 | Raf1 | 10310 | -0.400 | -0.1351 | Yes |
| 97 | Gp9 | 10360 | -0.405 | -0.1246 | Yes |
| 98 | Irf1 | 10434 | -0.415 | -0.1159 | Yes |
| 99 | Rbsn | 10930 | -0.513 | -0.1402 | Yes |
| 100 | Gnai3 | 11081 | -0.555 | -0.1331 | Yes |
| 101 | F5 | 11084 | -0.556 | -0.1130 | Yes |
| 102 | Dock9 | 11350 | -0.662 | -0.1119 | Yes |
| 103 | Dock10 | 11385 | -0.682 | -0.0901 | Yes |
| 104 | Gnb4 | 11403 | -0.693 | -0.0664 | Yes |
| 105 | Adam9 | 11441 | -0.727 | -0.0431 | Yes |
| 106 | Cebpb | 11495 | -0.789 | -0.0190 | Yes |
| 107 | Gca | 11501 | -0.793 | 0.0094 | Yes |