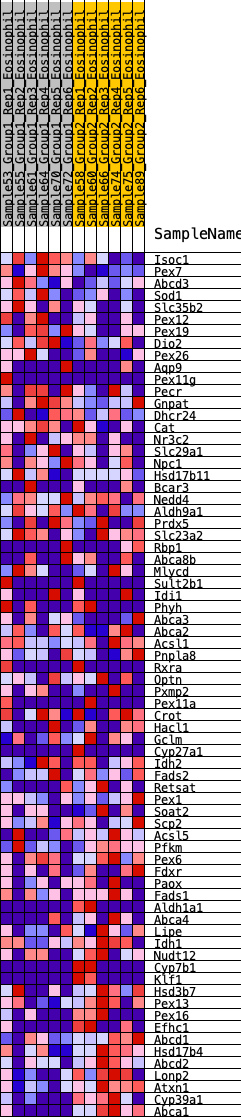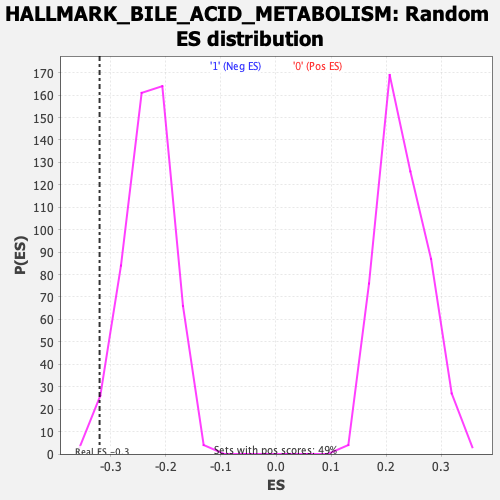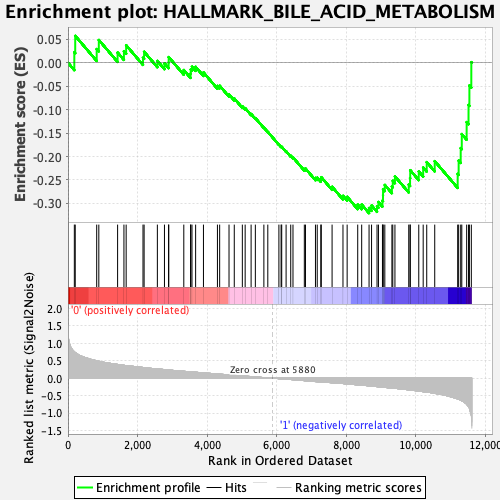
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.32073343 |
| Normalized Enrichment Score (NES) | -1.3803364 |
| Nominal p-value | 0.011811024 |
| FDR q-value | 0.21343394 |
| FWER p-Value | 0.57 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Isoc1 | 182 | 0.758 | 0.0224 | No |
| 2 | Pex7 | 208 | 0.735 | 0.0573 | No |
| 3 | Abcd3 | 825 | 0.501 | 0.0292 | No |
| 4 | Sod1 | 886 | 0.487 | 0.0485 | No |
| 5 | Slc35b2 | 1426 | 0.394 | 0.0217 | No |
| 6 | Pex12 | 1608 | 0.369 | 0.0246 | No |
| 7 | Pex19 | 1672 | 0.359 | 0.0372 | No |
| 8 | Dio2 | 2156 | 0.307 | 0.0109 | No |
| 9 | Pex26 | 2188 | 0.303 | 0.0235 | No |
| 10 | Aqp9 | 2569 | 0.266 | 0.0039 | No |
| 11 | Pex11g | 2773 | 0.247 | -0.0012 | No |
| 12 | Pecr | 2893 | 0.235 | 0.0003 | No |
| 13 | Gnpat | 2895 | 0.235 | 0.0121 | No |
| 14 | Dhcr24 | 3329 | 0.197 | -0.0155 | No |
| 15 | Cat | 3524 | 0.182 | -0.0232 | No |
| 16 | Nr3c2 | 3529 | 0.181 | -0.0144 | No |
| 17 | Slc29a1 | 3564 | 0.178 | -0.0083 | No |
| 18 | Npc1 | 3669 | 0.170 | -0.0088 | No |
| 19 | Hsd17b11 | 3894 | 0.150 | -0.0206 | No |
| 20 | Bcar3 | 4296 | 0.119 | -0.0494 | No |
| 21 | Nedd4 | 4360 | 0.114 | -0.0491 | No |
| 22 | Aldh9a1 | 4629 | 0.096 | -0.0674 | No |
| 23 | Prdx5 | 4780 | 0.084 | -0.0762 | No |
| 24 | Slc23a2 | 5012 | 0.065 | -0.0930 | No |
| 25 | Rbp1 | 5093 | 0.059 | -0.0969 | No |
| 26 | Abca8b | 5264 | 0.047 | -0.1093 | No |
| 27 | Mlycd | 5386 | 0.036 | -0.1180 | No |
| 28 | Sult2b1 | 5631 | 0.017 | -0.1382 | No |
| 29 | Idi1 | 5744 | 0.010 | -0.1474 | No |
| 30 | Phyh | 6063 | -0.012 | -0.1744 | No |
| 31 | Abca3 | 6126 | -0.018 | -0.1788 | No |
| 32 | Abca2 | 6144 | -0.020 | -0.1793 | No |
| 33 | Acsl1 | 6270 | -0.027 | -0.1888 | No |
| 34 | Pnpla8 | 6403 | -0.037 | -0.1984 | No |
| 35 | Rxra | 6468 | -0.041 | -0.2018 | No |
| 36 | Optn | 6794 | -0.069 | -0.2265 | No |
| 37 | Pxmp2 | 6827 | -0.072 | -0.2257 | No |
| 38 | Pex11a | 7114 | -0.096 | -0.2456 | No |
| 39 | Crot | 7165 | -0.099 | -0.2450 | No |
| 40 | Hacl1 | 7268 | -0.105 | -0.2485 | No |
| 41 | Gclm | 7281 | -0.106 | -0.2442 | No |
| 42 | Cyp27a1 | 7592 | -0.130 | -0.2645 | No |
| 43 | Idh2 | 7905 | -0.153 | -0.2838 | No |
| 44 | Fads2 | 8025 | -0.163 | -0.2859 | No |
| 45 | Retsat | 8329 | -0.190 | -0.3026 | No |
| 46 | Pex1 | 8443 | -0.198 | -0.3024 | No |
| 47 | Soat2 | 8655 | -0.219 | -0.3097 | Yes |
| 48 | Scp2 | 8727 | -0.226 | -0.3044 | Yes |
| 49 | Acsl5 | 8884 | -0.241 | -0.3058 | Yes |
| 50 | Pfkm | 8925 | -0.245 | -0.2969 | Yes |
| 51 | Pex6 | 9040 | -0.254 | -0.2940 | Yes |
| 52 | Fdxr | 9056 | -0.255 | -0.2824 | Yes |
| 53 | Paox | 9058 | -0.257 | -0.2696 | Yes |
| 54 | Fads1 | 9108 | -0.262 | -0.2606 | Yes |
| 55 | Aldh1a1 | 9312 | -0.283 | -0.2639 | Yes |
| 56 | Abca4 | 9337 | -0.284 | -0.2517 | Yes |
| 57 | Lipe | 9399 | -0.290 | -0.2424 | Yes |
| 58 | Idh1 | 9796 | -0.334 | -0.2598 | Yes |
| 59 | Nudt12 | 9834 | -0.339 | -0.2460 | Yes |
| 60 | Cyp7b1 | 9839 | -0.339 | -0.2292 | Yes |
| 61 | Klf1 | 10084 | -0.367 | -0.2319 | Yes |
| 62 | Hsd3b7 | 10211 | -0.386 | -0.2234 | Yes |
| 63 | Pex13 | 10313 | -0.400 | -0.2119 | Yes |
| 64 | Pex16 | 10543 | -0.435 | -0.2099 | Yes |
| 65 | Efhc1 | 11203 | -0.595 | -0.2370 | Yes |
| 66 | Abcd1 | 11231 | -0.607 | -0.2087 | Yes |
| 67 | Hsd17b4 | 11290 | -0.628 | -0.1820 | Yes |
| 68 | Abcd2 | 11321 | -0.646 | -0.1521 | Yes |
| 69 | Lonp2 | 11461 | -0.744 | -0.1266 | Yes |
| 70 | Atxn1 | 11515 | -0.816 | -0.0900 | Yes |
| 71 | Cyp39a1 | 11540 | -0.867 | -0.0484 | Yes |
| 72 | Abca1 | 11595 | -1.077 | 0.0012 | Yes |