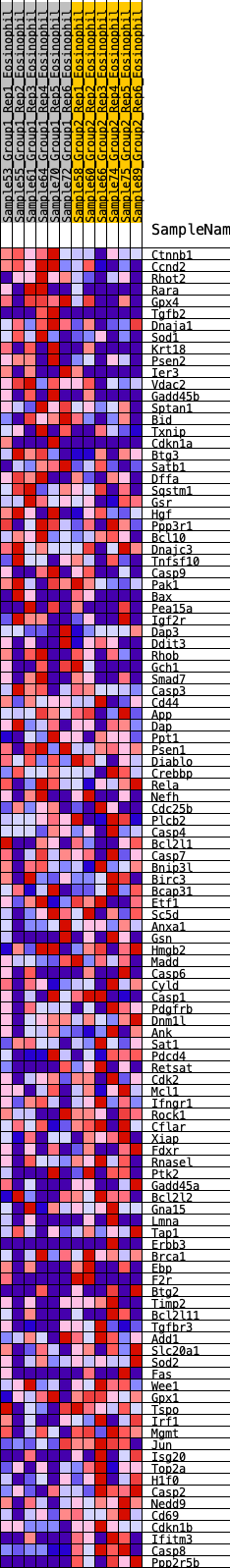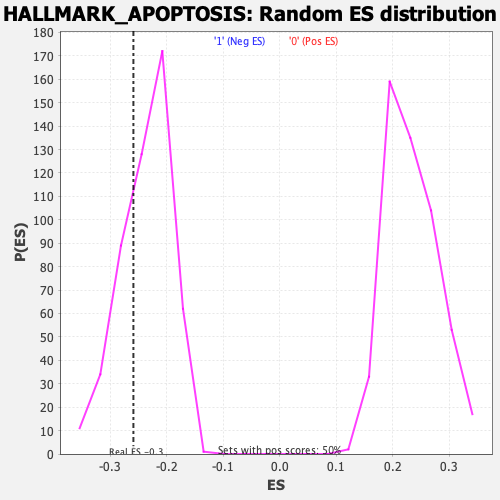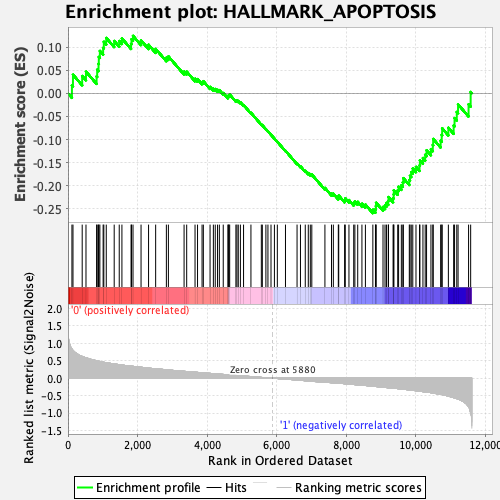
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.25931355 |
| Normalized Enrichment Score (NES) | -1.0918162 |
| Nominal p-value | 0.31187123 |
| FDR q-value | 0.47403395 |
| FWER p-Value | 0.989 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ctnnb1 | 113 | 0.837 | 0.0172 | No |
| 2 | Ccnd2 | 143 | 0.795 | 0.0403 | No |
| 3 | Rhot2 | 408 | 0.624 | 0.0374 | No |
| 4 | Rara | 517 | 0.583 | 0.0468 | No |
| 5 | Gpx4 | 822 | 0.502 | 0.0366 | No |
| 6 | Tgfb2 | 839 | 0.498 | 0.0512 | No |
| 7 | Dnaja1 | 878 | 0.489 | 0.0637 | No |
| 8 | Sod1 | 886 | 0.487 | 0.0788 | No |
| 9 | Krt18 | 916 | 0.478 | 0.0917 | No |
| 10 | Psen2 | 1010 | 0.460 | 0.0984 | No |
| 11 | Ier3 | 1030 | 0.455 | 0.1115 | No |
| 12 | Vdac2 | 1100 | 0.444 | 0.1198 | No |
| 13 | Gadd45b | 1328 | 0.410 | 0.1132 | No |
| 14 | Sptan1 | 1473 | 0.386 | 0.1132 | No |
| 15 | Bid | 1550 | 0.375 | 0.1187 | No |
| 16 | Txnip | 1813 | 0.344 | 0.1070 | No |
| 17 | Cdkn1a | 1826 | 0.343 | 0.1170 | No |
| 18 | Btg3 | 1869 | 0.339 | 0.1243 | No |
| 19 | Satb1 | 2100 | 0.314 | 0.1144 | No |
| 20 | Dffa | 2316 | 0.289 | 0.1050 | No |
| 21 | Sqstm1 | 2520 | 0.270 | 0.0961 | No |
| 22 | Gsr | 2828 | 0.242 | 0.0772 | No |
| 23 | Hgf | 2886 | 0.236 | 0.0798 | No |
| 24 | Ppp3r1 | 3337 | 0.197 | 0.0470 | No |
| 25 | Bcl10 | 3412 | 0.191 | 0.0467 | No |
| 26 | Dnajc3 | 3652 | 0.171 | 0.0315 | No |
| 27 | Tnfsf10 | 3724 | 0.166 | 0.0307 | No |
| 28 | Casp9 | 3854 | 0.154 | 0.0244 | No |
| 29 | Pak1 | 3891 | 0.150 | 0.0261 | No |
| 30 | Bax | 4084 | 0.137 | 0.0138 | No |
| 31 | Pea15a | 4179 | 0.130 | 0.0099 | No |
| 32 | Igf2r | 4233 | 0.125 | 0.0093 | No |
| 33 | Dap3 | 4299 | 0.119 | 0.0075 | No |
| 34 | Ddit3 | 4351 | 0.116 | 0.0068 | No |
| 35 | Rhob | 4463 | 0.108 | 0.0006 | No |
| 36 | Gch1 | 4598 | 0.098 | -0.0079 | No |
| 37 | Smad7 | 4609 | 0.097 | -0.0056 | No |
| 38 | Casp3 | 4625 | 0.096 | -0.0038 | No |
| 39 | Cd44 | 4648 | 0.094 | -0.0027 | No |
| 40 | App | 4827 | 0.079 | -0.0156 | No |
| 41 | Dap | 4857 | 0.077 | -0.0157 | No |
| 42 | Ppt1 | 4895 | 0.074 | -0.0165 | No |
| 43 | Psen1 | 4956 | 0.069 | -0.0195 | No |
| 44 | Diablo | 5047 | 0.063 | -0.0253 | No |
| 45 | Crebbp | 5259 | 0.047 | -0.0422 | No |
| 46 | Rela | 5558 | 0.022 | -0.0674 | No |
| 47 | Nefh | 5587 | 0.020 | -0.0692 | No |
| 48 | Cdc25b | 5689 | 0.014 | -0.0775 | No |
| 49 | Plcb2 | 5748 | 0.010 | -0.0822 | No |
| 50 | Casp4 | 5836 | 0.003 | -0.0897 | No |
| 51 | Bcl2l1 | 5934 | -0.002 | -0.0980 | No |
| 52 | Casp7 | 6018 | -0.009 | -0.1050 | No |
| 53 | Bnip3l | 6252 | -0.026 | -0.1244 | No |
| 54 | Birc3 | 6585 | -0.052 | -0.1516 | No |
| 55 | Bcap31 | 6683 | -0.060 | -0.1581 | No |
| 56 | Etf1 | 6821 | -0.071 | -0.1677 | No |
| 57 | Sc5d | 6908 | -0.079 | -0.1727 | No |
| 58 | Anxa1 | 6966 | -0.083 | -0.1749 | No |
| 59 | Gsn | 7009 | -0.088 | -0.1758 | No |
| 60 | Hmgb2 | 7386 | -0.115 | -0.2047 | No |
| 61 | Madd | 7575 | -0.129 | -0.2169 | No |
| 62 | Casp6 | 7628 | -0.131 | -0.2172 | No |
| 63 | Cyld | 7771 | -0.142 | -0.2250 | No |
| 64 | Casp1 | 7783 | -0.143 | -0.2213 | No |
| 65 | Pdgfrb | 7952 | -0.158 | -0.2309 | No |
| 66 | Dnm1l | 7971 | -0.159 | -0.2273 | No |
| 67 | Ank | 8077 | -0.167 | -0.2311 | No |
| 68 | Sat1 | 8214 | -0.180 | -0.2371 | No |
| 69 | Pdcd4 | 8247 | -0.183 | -0.2340 | No |
| 70 | Retsat | 8329 | -0.190 | -0.2349 | No |
| 71 | Cdk2 | 8450 | -0.199 | -0.2389 | No |
| 72 | Mcl1 | 8550 | -0.209 | -0.2408 | No |
| 73 | Ifngr1 | 8764 | -0.229 | -0.2519 | Yes |
| 74 | Rock1 | 8839 | -0.236 | -0.2508 | Yes |
| 75 | Cflar | 8851 | -0.237 | -0.2441 | Yes |
| 76 | Xiap | 8857 | -0.238 | -0.2368 | Yes |
| 77 | Fdxr | 9056 | -0.255 | -0.2458 | Yes |
| 78 | Rnasel | 9114 | -0.263 | -0.2423 | Yes |
| 79 | Ptk2 | 9155 | -0.268 | -0.2371 | Yes |
| 80 | Gadd45a | 9207 | -0.273 | -0.2328 | Yes |
| 81 | Bcl2l2 | 9214 | -0.273 | -0.2245 | Yes |
| 82 | Gna15 | 9346 | -0.285 | -0.2267 | Yes |
| 83 | Lmna | 9362 | -0.287 | -0.2188 | Yes |
| 84 | Tap1 | 9367 | -0.287 | -0.2099 | Yes |
| 85 | Erbb3 | 9479 | -0.299 | -0.2099 | Yes |
| 86 | Brca1 | 9504 | -0.301 | -0.2023 | Yes |
| 87 | Ebp | 9583 | -0.309 | -0.1991 | Yes |
| 88 | F2r | 9626 | -0.314 | -0.1926 | Yes |
| 89 | Btg2 | 9641 | -0.315 | -0.1837 | Yes |
| 90 | Timp2 | 9812 | -0.337 | -0.1876 | Yes |
| 91 | Bcl2l11 | 9832 | -0.338 | -0.1784 | Yes |
| 92 | Tgfbr3 | 9868 | -0.343 | -0.1703 | Yes |
| 93 | Add1 | 9912 | -0.348 | -0.1629 | Yes |
| 94 | Slc20a1 | 10004 | -0.358 | -0.1592 | Yes |
| 95 | Sod2 | 10106 | -0.370 | -0.1561 | Yes |
| 96 | Fas | 10117 | -0.371 | -0.1450 | Yes |
| 97 | Wee1 | 10204 | -0.385 | -0.1400 | Yes |
| 98 | Gpx1 | 10269 | -0.394 | -0.1329 | Yes |
| 99 | Tspo | 10308 | -0.400 | -0.1233 | Yes |
| 100 | Irf1 | 10434 | -0.415 | -0.1208 | Yes |
| 101 | Mgmt | 10488 | -0.426 | -0.1117 | Yes |
| 102 | Jun | 10499 | -0.428 | -0.0988 | Yes |
| 103 | Isg20 | 10713 | -0.463 | -0.1024 | Yes |
| 104 | Top2a | 10744 | -0.468 | -0.0899 | Yes |
| 105 | H1f0 | 10757 | -0.470 | -0.0758 | Yes |
| 106 | Casp2 | 10935 | -0.516 | -0.0745 | Yes |
| 107 | Nedd9 | 11087 | -0.557 | -0.0697 | Yes |
| 108 | Cd69 | 11113 | -0.566 | -0.0537 | Yes |
| 109 | Cdkn1b | 11177 | -0.584 | -0.0403 | Yes |
| 110 | Ifitm3 | 11214 | -0.602 | -0.0240 | Yes |
| 111 | Casp8 | 11516 | -0.817 | -0.0239 | Yes |
| 112 | Ppp2r5b | 11576 | -0.988 | 0.0029 | Yes |