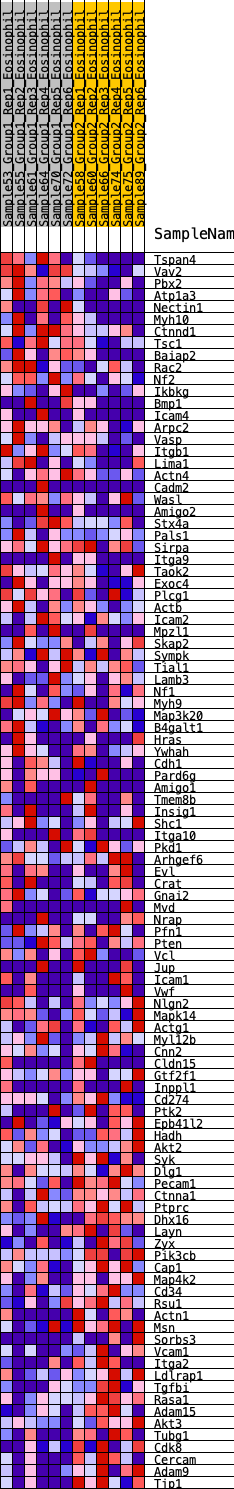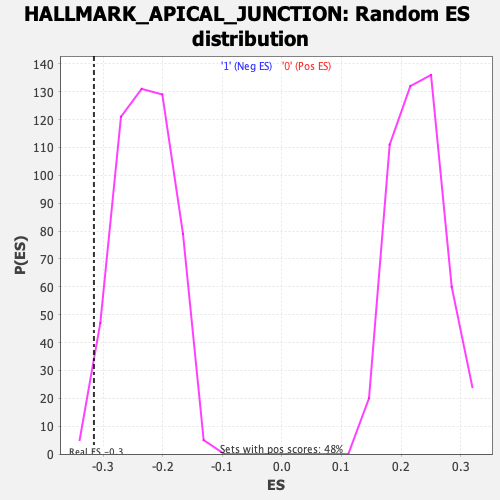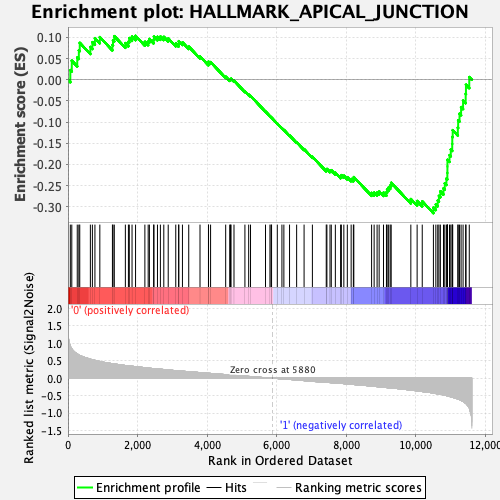
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group1_versus_Group2.Eosinophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Eosinophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | -0.31528145 |
| Normalized Enrichment Score (NES) | -1.3662388 |
| Nominal p-value | 0.023210831 |
| FDR q-value | 0.2014124 |
| FWER p-Value | 0.616 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tspan4 | 67 | 0.931 | 0.0228 | No |
| 2 | Vav2 | 109 | 0.841 | 0.0451 | No |
| 3 | Pbx2 | 266 | 0.693 | 0.0528 | No |
| 4 | Atp1a3 | 312 | 0.666 | 0.0693 | No |
| 5 | Nectin1 | 338 | 0.651 | 0.0872 | No |
| 6 | Myh10 | 643 | 0.545 | 0.0775 | No |
| 7 | Ctnnd1 | 701 | 0.529 | 0.0888 | No |
| 8 | Tsc1 | 774 | 0.512 | 0.0982 | No |
| 9 | Baiap2 | 915 | 0.478 | 0.1008 | No |
| 10 | Rac2 | 1277 | 0.416 | 0.0822 | No |
| 11 | Nf2 | 1294 | 0.414 | 0.0935 | No |
| 12 | Ikbkg | 1331 | 0.409 | 0.1030 | No |
| 13 | Bmp1 | 1649 | 0.363 | 0.0865 | No |
| 14 | Icam4 | 1737 | 0.353 | 0.0898 | No |
| 15 | Arpc2 | 1762 | 0.351 | 0.0985 | No |
| 16 | Vasp | 1841 | 0.342 | 0.1022 | No |
| 17 | Itgb1 | 1942 | 0.331 | 0.1037 | No |
| 18 | Lima1 | 2210 | 0.302 | 0.0898 | No |
| 19 | Actn4 | 2305 | 0.291 | 0.0905 | No |
| 20 | Cadm2 | 2340 | 0.287 | 0.0964 | No |
| 21 | Wasl | 2462 | 0.274 | 0.0943 | No |
| 22 | Amigo2 | 2471 | 0.273 | 0.1020 | No |
| 23 | Stx4a | 2572 | 0.266 | 0.1015 | No |
| 24 | Pals1 | 2656 | 0.257 | 0.1022 | No |
| 25 | Sirpa | 2754 | 0.248 | 0.1013 | No |
| 26 | Itga9 | 2881 | 0.236 | 0.0976 | No |
| 27 | Taok2 | 3098 | 0.218 | 0.0856 | No |
| 28 | Exoc4 | 3181 | 0.211 | 0.0849 | No |
| 29 | Plcg1 | 3185 | 0.210 | 0.0911 | No |
| 30 | Actb | 3290 | 0.201 | 0.0882 | No |
| 31 | Icam2 | 3471 | 0.186 | 0.0783 | No |
| 32 | Mpzl1 | 3795 | 0.159 | 0.0551 | No |
| 33 | Skap2 | 4038 | 0.139 | 0.0384 | No |
| 34 | Sympk | 4040 | 0.139 | 0.0426 | No |
| 35 | Tial1 | 4099 | 0.136 | 0.0417 | No |
| 36 | Lamb3 | 4533 | 0.103 | 0.0072 | No |
| 37 | Nf1 | 4649 | 0.094 | 0.0001 | No |
| 38 | Myh9 | 4672 | 0.092 | 0.0011 | No |
| 39 | Map3k20 | 4687 | 0.091 | 0.0026 | No |
| 40 | B4galt1 | 4772 | 0.085 | -0.0020 | No |
| 41 | Hras | 5086 | 0.059 | -0.0274 | No |
| 42 | Ywhah | 5194 | 0.051 | -0.0352 | No |
| 43 | Cdh1 | 5247 | 0.047 | -0.0382 | No |
| 44 | Pard6g | 5678 | 0.015 | -0.0751 | No |
| 45 | Amigo1 | 5806 | 0.006 | -0.0860 | No |
| 46 | Tmem8b | 5839 | 0.003 | -0.0887 | No |
| 47 | Insig1 | 5852 | 0.002 | -0.0896 | No |
| 48 | Shc1 | 6017 | -0.009 | -0.1036 | No |
| 49 | Itga10 | 6150 | -0.020 | -0.1145 | No |
| 50 | Pkd1 | 6206 | -0.023 | -0.1186 | No |
| 51 | Arhgef6 | 6369 | -0.034 | -0.1316 | No |
| 52 | Evl | 6573 | -0.051 | -0.1477 | No |
| 53 | Crat | 6787 | -0.069 | -0.1641 | No |
| 54 | Gnai2 | 7026 | -0.088 | -0.1821 | No |
| 55 | Mvd | 7424 | -0.118 | -0.2129 | No |
| 56 | Nrap | 7443 | -0.118 | -0.2109 | No |
| 57 | Pfn1 | 7532 | -0.125 | -0.2147 | No |
| 58 | Pten | 7569 | -0.128 | -0.2139 | No |
| 59 | Vcl | 7686 | -0.136 | -0.2198 | No |
| 60 | Jup | 7837 | -0.148 | -0.2283 | No |
| 61 | Icam1 | 7859 | -0.149 | -0.2255 | No |
| 62 | Vwf | 7930 | -0.155 | -0.2268 | No |
| 63 | Nlgn2 | 8030 | -0.163 | -0.2304 | No |
| 64 | Mapk14 | 8140 | -0.172 | -0.2346 | No |
| 65 | Actg1 | 8204 | -0.179 | -0.2346 | No |
| 66 | Myl12b | 8217 | -0.180 | -0.2301 | No |
| 67 | Cnn2 | 8728 | -0.226 | -0.2675 | No |
| 68 | Cldn15 | 8801 | -0.233 | -0.2666 | No |
| 69 | Gtf2f1 | 8886 | -0.241 | -0.2665 | No |
| 70 | Inppl1 | 8945 | -0.247 | -0.2639 | No |
| 71 | Cd274 | 9070 | -0.258 | -0.2668 | No |
| 72 | Ptk2 | 9155 | -0.268 | -0.2659 | No |
| 73 | Epb41l2 | 9174 | -0.270 | -0.2591 | No |
| 74 | Hadh | 9218 | -0.274 | -0.2545 | No |
| 75 | Akt2 | 9263 | -0.278 | -0.2498 | No |
| 76 | Syk | 9291 | -0.281 | -0.2435 | No |
| 77 | Dlg1 | 9856 | -0.341 | -0.2820 | No |
| 78 | Pecam1 | 10037 | -0.362 | -0.2865 | No |
| 79 | Ctnna1 | 10184 | -0.382 | -0.2875 | No |
| 80 | Ptprc | 10505 | -0.429 | -0.3021 | Yes |
| 81 | Dhx16 | 10575 | -0.440 | -0.2946 | Yes |
| 82 | Layn | 10630 | -0.449 | -0.2855 | Yes |
| 83 | Zyx | 10664 | -0.455 | -0.2744 | Yes |
| 84 | Pik3cb | 10705 | -0.461 | -0.2637 | Yes |
| 85 | Cap1 | 10795 | -0.480 | -0.2567 | Yes |
| 86 | Map4k2 | 10832 | -0.489 | -0.2448 | Yes |
| 87 | Cd34 | 10879 | -0.499 | -0.2335 | Yes |
| 88 | Rsu1 | 10906 | -0.505 | -0.2202 | Yes |
| 89 | Actn1 | 10908 | -0.505 | -0.2048 | Yes |
| 90 | Msn | 10909 | -0.506 | -0.1893 | Yes |
| 91 | Sorbs3 | 10968 | -0.523 | -0.1782 | Yes |
| 92 | Vcam1 | 11005 | -0.534 | -0.1650 | Yes |
| 93 | Itga2 | 11046 | -0.544 | -0.1517 | Yes |
| 94 | Ldlrap1 | 11048 | -0.545 | -0.1351 | Yes |
| 95 | Tgfbi | 11060 | -0.549 | -0.1192 | Yes |
| 96 | Rasa1 | 11207 | -0.599 | -0.1135 | Yes |
| 97 | Adam15 | 11217 | -0.602 | -0.0958 | Yes |
| 98 | Akt3 | 11256 | -0.614 | -0.0802 | Yes |
| 99 | Tubg1 | 11301 | -0.632 | -0.0646 | Yes |
| 100 | Cdk8 | 11358 | -0.665 | -0.0491 | Yes |
| 101 | Cercam | 11433 | -0.719 | -0.0334 | Yes |
| 102 | Adam9 | 11441 | -0.727 | -0.0117 | Yes |
| 103 | Tjp1 | 11536 | -0.854 | 0.0063 | Yes |