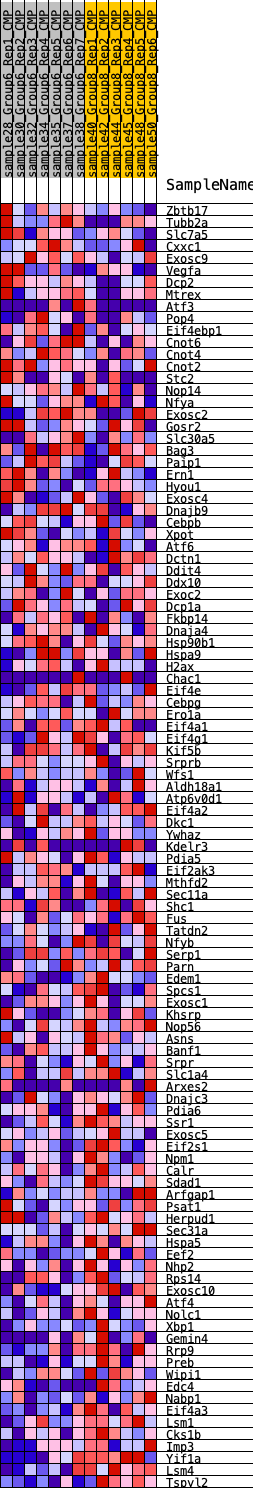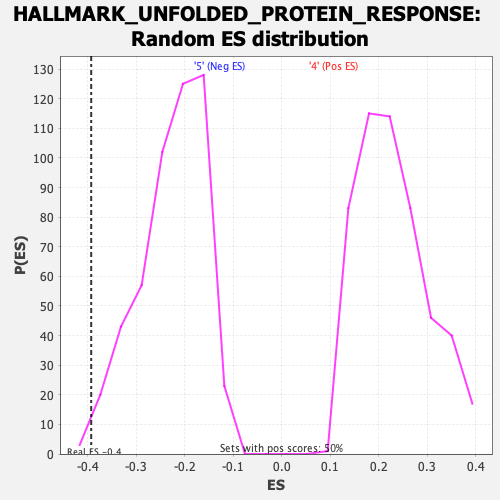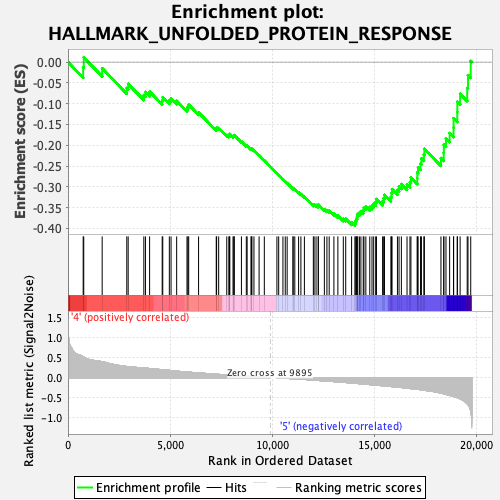
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group6_versus_Group8.CMP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | CMP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.3932773 |
| Normalized Enrichment Score (NES) | -1.7390169 |
| Nominal p-value | 0.007984032 |
| FDR q-value | 0.15319903 |
| FWER p-Value | 0.091 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Zbtb17 | 743 | 0.515 | -0.0124 | No |
| 2 | Tubb2a | 771 | 0.506 | 0.0112 | No |
| 3 | Slc7a5 | 1673 | 0.399 | -0.0149 | No |
| 4 | Cxxc1 | 2878 | 0.278 | -0.0625 | No |
| 5 | Exosc9 | 2946 | 0.273 | -0.0524 | No |
| 6 | Vegfa | 3713 | 0.239 | -0.0795 | No |
| 7 | Dcp2 | 3792 | 0.235 | -0.0719 | No |
| 8 | Mtrex | 3996 | 0.227 | -0.0711 | No |
| 9 | Atf3 | 4611 | 0.196 | -0.0926 | No |
| 10 | Pop4 | 4640 | 0.195 | -0.0844 | No |
| 11 | Eif4ebp1 | 4961 | 0.180 | -0.0918 | No |
| 12 | Cnot6 | 5045 | 0.176 | -0.0874 | No |
| 13 | Cnot4 | 5320 | 0.162 | -0.0933 | No |
| 14 | Cnot2 | 5826 | 0.141 | -0.1120 | No |
| 15 | Stc2 | 5869 | 0.139 | -0.1073 | No |
| 16 | Nop14 | 5910 | 0.138 | -0.1025 | No |
| 17 | Nfya | 6393 | 0.118 | -0.1213 | No |
| 18 | Exosc2 | 7258 | 0.086 | -0.1610 | No |
| 19 | Gosr2 | 7276 | 0.085 | -0.1576 | No |
| 20 | Slc30a5 | 7377 | 0.082 | -0.1587 | No |
| 21 | Bag3 | 7773 | 0.069 | -0.1754 | No |
| 22 | Paip1 | 7874 | 0.066 | -0.1772 | No |
| 23 | Ern1 | 7915 | 0.065 | -0.1760 | No |
| 24 | Hyou1 | 7917 | 0.065 | -0.1729 | No |
| 25 | Exosc4 | 8080 | 0.059 | -0.1782 | No |
| 26 | Dnajb9 | 8115 | 0.058 | -0.1771 | No |
| 27 | Cebpb | 8144 | 0.057 | -0.1757 | No |
| 28 | Xpot | 8491 | 0.047 | -0.1910 | No |
| 29 | Atf6 | 8730 | 0.038 | -0.2012 | No |
| 30 | Dctn1 | 8770 | 0.037 | -0.2014 | No |
| 31 | Ddit4 | 8958 | 0.031 | -0.2093 | No |
| 32 | Ddx10 | 8991 | 0.030 | -0.2095 | No |
| 33 | Exoc2 | 9001 | 0.030 | -0.2085 | No |
| 34 | Dcp1a | 9104 | 0.026 | -0.2124 | No |
| 35 | Fkbp14 | 9356 | 0.018 | -0.2243 | No |
| 36 | Dnaja4 | 9612 | 0.010 | -0.2367 | No |
| 37 | Hsp90b1 | 10229 | -0.001 | -0.2680 | No |
| 38 | Hspa9 | 10315 | -0.003 | -0.2722 | No |
| 39 | H2ax | 10523 | -0.010 | -0.2822 | No |
| 40 | Chac1 | 10637 | -0.014 | -0.2873 | No |
| 41 | Eif4e | 10730 | -0.016 | -0.2912 | No |
| 42 | Cebpg | 11016 | -0.026 | -0.3044 | No |
| 43 | Ero1a | 11021 | -0.026 | -0.3033 | No |
| 44 | Eif4a1 | 11104 | -0.029 | -0.3060 | No |
| 45 | Eif4g1 | 11290 | -0.035 | -0.3137 | No |
| 46 | Kif5b | 11403 | -0.039 | -0.3175 | No |
| 47 | Srprb | 11574 | -0.043 | -0.3240 | No |
| 48 | Wfs1 | 12012 | -0.059 | -0.3433 | No |
| 49 | Aldh18a1 | 12049 | -0.060 | -0.3422 | No |
| 50 | Atp6v0d1 | 12144 | -0.063 | -0.3438 | No |
| 51 | Eif4a2 | 12252 | -0.068 | -0.3459 | No |
| 52 | Dkc1 | 12255 | -0.068 | -0.3427 | No |
| 53 | Ywhaz | 12552 | -0.079 | -0.3539 | No |
| 54 | Kdelr3 | 12677 | -0.083 | -0.3561 | No |
| 55 | Pdia5 | 12788 | -0.086 | -0.3574 | No |
| 56 | Eif2ak3 | 13019 | -0.096 | -0.3644 | No |
| 57 | Mthfd2 | 13213 | -0.104 | -0.3691 | No |
| 58 | Sec11a | 13479 | -0.115 | -0.3769 | No |
| 59 | Shc1 | 13597 | -0.119 | -0.3770 | No |
| 60 | Fus | 13885 | -0.131 | -0.3851 | No |
| 61 | Tatdn2 | 14047 | -0.138 | -0.3865 | Yes |
| 62 | Nfyb | 14088 | -0.140 | -0.3816 | Yes |
| 63 | Serp1 | 14123 | -0.141 | -0.3764 | Yes |
| 64 | Parn | 14157 | -0.142 | -0.3710 | Yes |
| 65 | Edem1 | 14185 | -0.144 | -0.3653 | Yes |
| 66 | Spcs1 | 14268 | -0.147 | -0.3622 | Yes |
| 67 | Exosc1 | 14341 | -0.151 | -0.3584 | Yes |
| 68 | Khsrp | 14455 | -0.155 | -0.3565 | Yes |
| 69 | Nop56 | 14490 | -0.157 | -0.3505 | Yes |
| 70 | Asns | 14583 | -0.161 | -0.3473 | Yes |
| 71 | Banf1 | 14775 | -0.170 | -0.3486 | Yes |
| 72 | Srpr | 14884 | -0.175 | -0.3455 | Yes |
| 73 | Slc1a4 | 14966 | -0.179 | -0.3407 | Yes |
| 74 | Arxes2 | 15064 | -0.184 | -0.3366 | Yes |
| 75 | Dnajc3 | 15103 | -0.186 | -0.3294 | Yes |
| 76 | Pdia6 | 15403 | -0.199 | -0.3348 | Yes |
| 77 | Ssr1 | 15456 | -0.201 | -0.3275 | Yes |
| 78 | Exosc5 | 15495 | -0.203 | -0.3194 | Yes |
| 79 | Eif2s1 | 15809 | -0.218 | -0.3246 | Yes |
| 80 | Npm1 | 15839 | -0.219 | -0.3152 | Yes |
| 81 | Calr | 15865 | -0.221 | -0.3056 | Yes |
| 82 | Sdad1 | 16130 | -0.235 | -0.3074 | Yes |
| 83 | Arfgap1 | 16208 | -0.240 | -0.2995 | Yes |
| 84 | Psat1 | 16322 | -0.242 | -0.2933 | Yes |
| 85 | Herpud1 | 16599 | -0.258 | -0.2946 | Yes |
| 86 | Sec31a | 16742 | -0.267 | -0.2887 | Yes |
| 87 | Hspa5 | 16790 | -0.270 | -0.2777 | Yes |
| 88 | Eef2 | 17101 | -0.286 | -0.2794 | Yes |
| 89 | Nhp2 | 17103 | -0.286 | -0.2654 | Yes |
| 90 | Rps14 | 17141 | -0.289 | -0.2530 | Yes |
| 91 | Exosc10 | 17261 | -0.296 | -0.2444 | Yes |
| 92 | Atf4 | 17304 | -0.300 | -0.2318 | Yes |
| 93 | Nolc1 | 17424 | -0.309 | -0.2226 | Yes |
| 94 | Xbp1 | 17449 | -0.311 | -0.2085 | Yes |
| 95 | Gemin4 | 18260 | -0.381 | -0.2309 | Yes |
| 96 | Rrp9 | 18396 | -0.398 | -0.2181 | Yes |
| 97 | Preb | 18402 | -0.399 | -0.1987 | Yes |
| 98 | Wipi1 | 18506 | -0.414 | -0.1835 | Yes |
| 99 | Edc4 | 18685 | -0.441 | -0.1708 | Yes |
| 100 | Nabp1 | 18877 | -0.463 | -0.1577 | Yes |
| 101 | Eif4a3 | 18880 | -0.464 | -0.1350 | Yes |
| 102 | Lsm1 | 19060 | -0.494 | -0.1197 | Yes |
| 103 | Cks1b | 19063 | -0.495 | -0.0954 | Yes |
| 104 | Imp3 | 19199 | -0.524 | -0.0764 | Yes |
| 105 | Yif1a | 19545 | -0.642 | -0.0623 | Yes |
| 106 | Lsm4 | 19584 | -0.665 | -0.0314 | Yes |
| 107 | Tspyl2 | 19718 | -0.829 | 0.0027 | Yes |