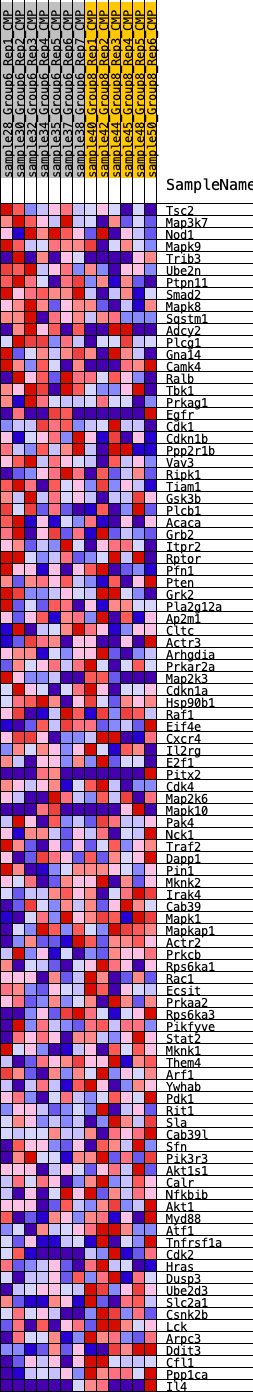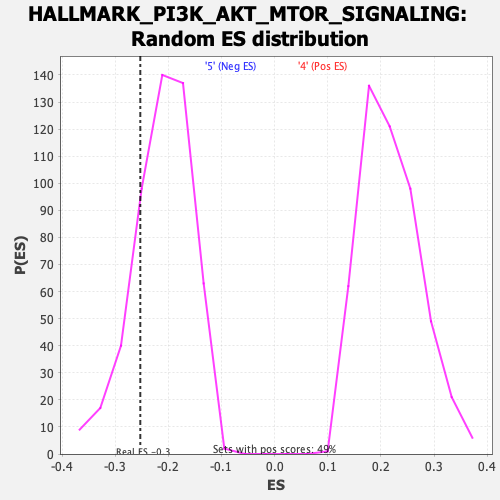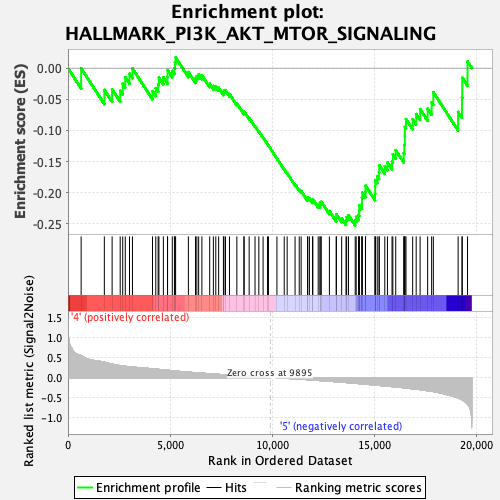
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group6_versus_Group8.CMP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | CMP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.25273842 |
| Normalized Enrichment Score (NES) | -1.1996304 |
| Nominal p-value | 0.21343873 |
| FDR q-value | 0.29759857 |
| FWER p-Value | 0.905 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tsc2 | 643 | 0.545 | -0.0001 | No |
| 2 | Map3k7 | 1779 | 0.384 | -0.0349 | No |
| 3 | Nod1 | 2160 | 0.337 | -0.0341 | No |
| 4 | Mapk9 | 2555 | 0.300 | -0.0362 | No |
| 5 | Trib3 | 2679 | 0.293 | -0.0250 | No |
| 6 | Ube2n | 2804 | 0.283 | -0.0144 | No |
| 7 | Ptpn11 | 3012 | 0.269 | -0.0089 | No |
| 8 | Smad2 | 3156 | 0.263 | -0.0004 | No |
| 9 | Mapk8 | 4138 | 0.224 | -0.0369 | No |
| 10 | Sqstm1 | 4298 | 0.215 | -0.0322 | No |
| 11 | Adcy2 | 4413 | 0.207 | -0.0256 | No |
| 12 | Plcg1 | 4451 | 0.205 | -0.0152 | No |
| 13 | Gna14 | 4670 | 0.194 | -0.0147 | No |
| 14 | Camk4 | 4872 | 0.184 | -0.0139 | No |
| 15 | Ralb | 4875 | 0.184 | -0.0030 | No |
| 16 | Tbk1 | 5106 | 0.173 | -0.0044 | No |
| 17 | Prkag1 | 5210 | 0.167 | 0.0004 | No |
| 18 | Egfr | 5238 | 0.166 | 0.0089 | No |
| 19 | Cdk1 | 5269 | 0.165 | 0.0172 | No |
| 20 | Cdkn1b | 5894 | 0.138 | -0.0063 | No |
| 21 | Ppp2r1b | 6242 | 0.123 | -0.0165 | No |
| 22 | Vav3 | 6309 | 0.121 | -0.0127 | No |
| 23 | Ripk1 | 6397 | 0.117 | -0.0101 | No |
| 24 | Tiam1 | 6555 | 0.112 | -0.0114 | No |
| 25 | Gsk3b | 6938 | 0.098 | -0.0250 | No |
| 26 | Plcb1 | 7117 | 0.092 | -0.0285 | No |
| 27 | Acaca | 7236 | 0.086 | -0.0294 | No |
| 28 | Grb2 | 7375 | 0.082 | -0.0315 | No |
| 29 | Itpr2 | 7609 | 0.074 | -0.0390 | No |
| 30 | Rptor | 7636 | 0.073 | -0.0359 | No |
| 31 | Pfn1 | 7710 | 0.071 | -0.0354 | No |
| 32 | Pten | 7906 | 0.065 | -0.0415 | No |
| 33 | Grk2 | 8267 | 0.053 | -0.0566 | No |
| 34 | Pla2g12a | 8614 | 0.043 | -0.0716 | No |
| 35 | Ap2m1 | 8622 | 0.043 | -0.0694 | No |
| 36 | Cltc | 8870 | 0.034 | -0.0800 | No |
| 37 | Actr3 | 9157 | 0.025 | -0.0930 | No |
| 38 | Arhgdia | 9343 | 0.018 | -0.1014 | No |
| 39 | Prkar2a | 9552 | 0.012 | -0.1113 | No |
| 40 | Map2k3 | 9766 | 0.004 | -0.1218 | No |
| 41 | Cdkn1a | 9815 | 0.003 | -0.1241 | No |
| 42 | Hsp90b1 | 10229 | -0.001 | -0.1451 | No |
| 43 | Raf1 | 10587 | -0.012 | -0.1625 | No |
| 44 | Eif4e | 10730 | -0.016 | -0.1687 | No |
| 45 | Cxcr4 | 11117 | -0.029 | -0.1866 | No |
| 46 | Il2rg | 11322 | -0.036 | -0.1948 | No |
| 47 | E2f1 | 11415 | -0.039 | -0.1972 | No |
| 48 | Pitx2 | 11730 | -0.048 | -0.2103 | No |
| 49 | Cdk4 | 11737 | -0.048 | -0.2077 | No |
| 50 | Map2k6 | 11819 | -0.051 | -0.2088 | No |
| 51 | Mapk10 | 11972 | -0.058 | -0.2131 | No |
| 52 | Pak4 | 11987 | -0.058 | -0.2103 | No |
| 53 | Nck1 | 12250 | -0.068 | -0.2196 | No |
| 54 | Traf2 | 12341 | -0.071 | -0.2199 | No |
| 55 | Dapp1 | 12342 | -0.071 | -0.2157 | No |
| 56 | Pin1 | 12401 | -0.073 | -0.2143 | No |
| 57 | Mknk2 | 12798 | -0.087 | -0.2292 | No |
| 58 | Irak4 | 13133 | -0.100 | -0.2402 | No |
| 59 | Cab39 | 13139 | -0.101 | -0.2345 | No |
| 60 | Mapk1 | 13399 | -0.111 | -0.2410 | No |
| 61 | Mapkap1 | 13612 | -0.119 | -0.2446 | Yes |
| 62 | Actr2 | 13642 | -0.120 | -0.2389 | Yes |
| 63 | Prkcb | 13732 | -0.125 | -0.2360 | Yes |
| 64 | Rps6ka1 | 14062 | -0.139 | -0.2445 | Yes |
| 65 | Rac1 | 14110 | -0.141 | -0.2385 | Yes |
| 66 | Ecsit | 14225 | -0.145 | -0.2356 | Yes |
| 67 | Prkaa2 | 14259 | -0.147 | -0.2285 | Yes |
| 68 | Rps6ka3 | 14266 | -0.147 | -0.2200 | Yes |
| 69 | Pikfyve | 14390 | -0.153 | -0.2171 | Yes |
| 70 | Stat2 | 14392 | -0.153 | -0.2081 | Yes |
| 71 | Mknk1 | 14408 | -0.153 | -0.1997 | Yes |
| 72 | Them4 | 14563 | -0.160 | -0.1980 | Yes |
| 73 | Arf1 | 14568 | -0.160 | -0.1886 | Yes |
| 74 | Ywhab | 15033 | -0.183 | -0.2013 | Yes |
| 75 | Pdk1 | 15041 | -0.183 | -0.1908 | Yes |
| 76 | Rit1 | 15043 | -0.183 | -0.1799 | Yes |
| 77 | Sla | 15140 | -0.187 | -0.1736 | Yes |
| 78 | Cab39l | 15230 | -0.190 | -0.1668 | Yes |
| 79 | Sfn | 15241 | -0.191 | -0.1559 | Yes |
| 80 | Pik3r3 | 15511 | -0.204 | -0.1574 | Yes |
| 81 | Akt1s1 | 15641 | -0.210 | -0.1514 | Yes |
| 82 | Calr | 15865 | -0.221 | -0.1496 | Yes |
| 83 | Nfkbib | 15905 | -0.222 | -0.1383 | Yes |
| 84 | Akt1 | 16046 | -0.231 | -0.1316 | Yes |
| 85 | Myd88 | 16436 | -0.249 | -0.1365 | Yes |
| 86 | Atf1 | 16466 | -0.251 | -0.1230 | Yes |
| 87 | Tnfrsf1a | 16491 | -0.252 | -0.1092 | Yes |
| 88 | Cdk2 | 16494 | -0.252 | -0.0943 | Yes |
| 89 | Hras | 16548 | -0.255 | -0.0817 | Yes |
| 90 | Dusp3 | 16875 | -0.275 | -0.0818 | Yes |
| 91 | Ube2d3 | 17047 | -0.284 | -0.0736 | Yes |
| 92 | Slc2a1 | 17239 | -0.295 | -0.0657 | Yes |
| 93 | Csnk2b | 17608 | -0.324 | -0.0650 | Yes |
| 94 | Lck | 17804 | -0.336 | -0.0549 | Yes |
| 95 | Arpc3 | 17887 | -0.343 | -0.0385 | Yes |
| 96 | Ddit3 | 19101 | -0.502 | -0.0702 | Yes |
| 97 | Cfl1 | 19290 | -0.544 | -0.0473 | Yes |
| 98 | Ppp1ca | 19304 | -0.547 | -0.0153 | Yes |
| 99 | Il4 | 19561 | -0.653 | 0.0107 | Yes |