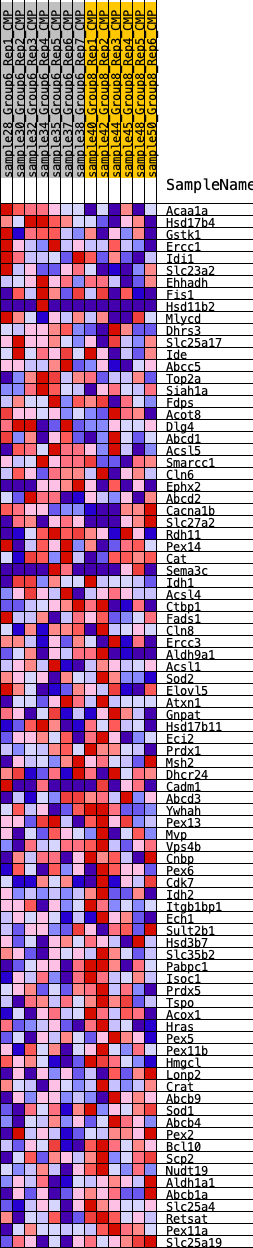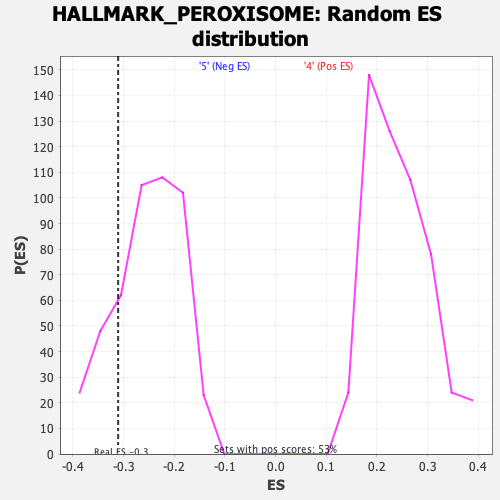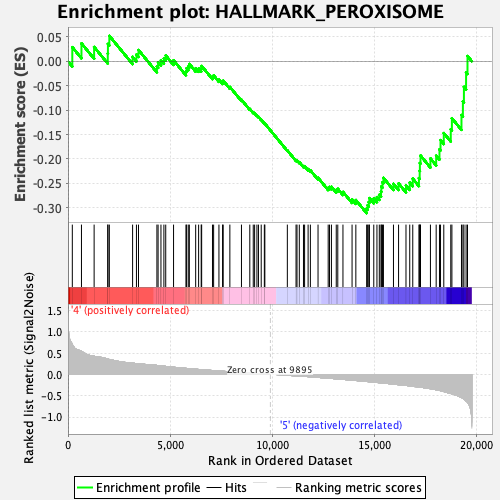
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group6_versus_Group8.CMP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | CMP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.3108408 |
| Normalized Enrichment Score (NES) | -1.2327573 |
| Nominal p-value | 0.19067797 |
| FDR q-value | 0.29946175 |
| FWER p-Value | 0.862 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Acaa1a | 211 | 0.701 | 0.0288 | No |
| 2 | Hsd17b4 | 659 | 0.542 | 0.0367 | No |
| 3 | Gstk1 | 1279 | 0.427 | 0.0293 | No |
| 4 | Ercc1 | 1947 | 0.361 | 0.0157 | No |
| 5 | Idi1 | 1955 | 0.360 | 0.0357 | No |
| 6 | Slc23a2 | 2024 | 0.351 | 0.0520 | No |
| 7 | Ehhadh | 3163 | 0.263 | 0.0090 | No |
| 8 | Fis1 | 3351 | 0.254 | 0.0139 | No |
| 9 | Hsd11b2 | 3451 | 0.252 | 0.0230 | No |
| 10 | Mlycd | 4353 | 0.212 | -0.0108 | No |
| 11 | Dhrs3 | 4411 | 0.208 | -0.0020 | No |
| 12 | Slc25a17 | 4551 | 0.199 | 0.0022 | No |
| 13 | Ide | 4695 | 0.193 | 0.0058 | No |
| 14 | Abcc5 | 4785 | 0.189 | 0.0119 | No |
| 15 | Top2a | 5168 | 0.170 | 0.0021 | No |
| 16 | Siah1a | 5775 | 0.143 | -0.0207 | No |
| 17 | Fdps | 5807 | 0.142 | -0.0143 | No |
| 18 | Acot8 | 5888 | 0.138 | -0.0105 | No |
| 19 | Dlg4 | 5941 | 0.136 | -0.0055 | No |
| 20 | Abcd1 | 6251 | 0.123 | -0.0142 | No |
| 21 | Acsl5 | 6395 | 0.118 | -0.0149 | No |
| 22 | Smarcc1 | 6521 | 0.113 | -0.0149 | No |
| 23 | Cln6 | 6544 | 0.112 | -0.0096 | No |
| 24 | Ephx2 | 7074 | 0.093 | -0.0313 | No |
| 25 | Abcd2 | 7130 | 0.091 | -0.0290 | No |
| 26 | Cacna1b | 7390 | 0.081 | -0.0375 | No |
| 27 | Slc27a2 | 7567 | 0.075 | -0.0422 | No |
| 28 | Rdh11 | 7598 | 0.074 | -0.0396 | No |
| 29 | Pex14 | 7925 | 0.064 | -0.0525 | No |
| 30 | Cat | 8493 | 0.046 | -0.0787 | No |
| 31 | Sema3c | 8901 | 0.033 | -0.0975 | No |
| 32 | Idh1 | 9070 | 0.027 | -0.1045 | No |
| 33 | Acsl4 | 9137 | 0.025 | -0.1064 | No |
| 34 | Ctbp1 | 9250 | 0.021 | -0.1110 | No |
| 35 | Fads1 | 9325 | 0.018 | -0.1137 | No |
| 36 | Cln8 | 9460 | 0.014 | -0.1197 | No |
| 37 | Ercc3 | 9613 | 0.010 | -0.1268 | No |
| 38 | Aldh9a1 | 9638 | 0.009 | -0.1276 | No |
| 39 | Acsl1 | 10739 | -0.016 | -0.1825 | No |
| 40 | Sod2 | 11165 | -0.031 | -0.2023 | No |
| 41 | Elovl5 | 11212 | -0.033 | -0.2028 | No |
| 42 | Atxn1 | 11325 | -0.036 | -0.2065 | No |
| 43 | Gnpat | 11529 | -0.042 | -0.2144 | No |
| 44 | Hsd17b11 | 11584 | -0.043 | -0.2147 | No |
| 45 | Eci2 | 11756 | -0.049 | -0.2207 | No |
| 46 | Prdx1 | 11867 | -0.053 | -0.2233 | No |
| 47 | Msh2 | 12242 | -0.068 | -0.2385 | No |
| 48 | Dhcr24 | 12739 | -0.085 | -0.2589 | No |
| 49 | Cadm1 | 12804 | -0.087 | -0.2572 | No |
| 50 | Abcd3 | 12898 | -0.090 | -0.2568 | No |
| 51 | Ywhah | 13134 | -0.100 | -0.2631 | No |
| 52 | Pex13 | 13209 | -0.104 | -0.2610 | No |
| 53 | Mvp | 13462 | -0.114 | -0.2674 | No |
| 54 | Vps4b | 13908 | -0.132 | -0.2826 | No |
| 55 | Cnbp | 14093 | -0.140 | -0.2840 | No |
| 56 | Pex6 | 14622 | -0.162 | -0.3017 | Yes |
| 57 | Cdk7 | 14670 | -0.165 | -0.2948 | Yes |
| 58 | Idh2 | 14727 | -0.168 | -0.2881 | Yes |
| 59 | Itgb1bp1 | 14758 | -0.170 | -0.2801 | Yes |
| 60 | Ech1 | 14971 | -0.180 | -0.2807 | Yes |
| 61 | Sult2b1 | 15126 | -0.186 | -0.2781 | Yes |
| 62 | Hsd3b7 | 15242 | -0.191 | -0.2731 | Yes |
| 63 | Slc35b2 | 15326 | -0.196 | -0.2663 | Yes |
| 64 | Pabpc1 | 15343 | -0.196 | -0.2561 | Yes |
| 65 | Isoc1 | 15397 | -0.199 | -0.2476 | Yes |
| 66 | Prdx5 | 15444 | -0.201 | -0.2386 | Yes |
| 67 | Tspo | 15935 | -0.224 | -0.2509 | Yes |
| 68 | Acox1 | 16185 | -0.239 | -0.2501 | Yes |
| 69 | Hras | 16548 | -0.255 | -0.2541 | Yes |
| 70 | Pex5 | 16732 | -0.266 | -0.2483 | Yes |
| 71 | Pex11b | 16880 | -0.276 | -0.2403 | Yes |
| 72 | Hmgcl | 17182 | -0.292 | -0.2391 | Yes |
| 73 | Lonp2 | 17219 | -0.294 | -0.2243 | Yes |
| 74 | Crat | 17229 | -0.295 | -0.2082 | Yes |
| 75 | Abcb9 | 17259 | -0.296 | -0.1929 | Yes |
| 76 | Sod1 | 17745 | -0.331 | -0.1989 | Yes |
| 77 | Abcb4 | 18026 | -0.357 | -0.1930 | Yes |
| 78 | Pex2 | 18188 | -0.373 | -0.1802 | Yes |
| 79 | Bcl10 | 18239 | -0.379 | -0.1614 | Yes |
| 80 | Scp2 | 18399 | -0.399 | -0.1470 | Yes |
| 81 | Nudt19 | 18741 | -0.446 | -0.1392 | Yes |
| 82 | Aldh1a1 | 18796 | -0.449 | -0.1166 | Yes |
| 83 | Abcb1a | 19265 | -0.538 | -0.1100 | Yes |
| 84 | Slc25a4 | 19335 | -0.556 | -0.0822 | Yes |
| 85 | Retsat | 19385 | -0.574 | -0.0523 | Yes |
| 86 | Pex11a | 19491 | -0.621 | -0.0226 | Yes |
| 87 | Slc25a19 | 19560 | -0.652 | 0.0107 | Yes |