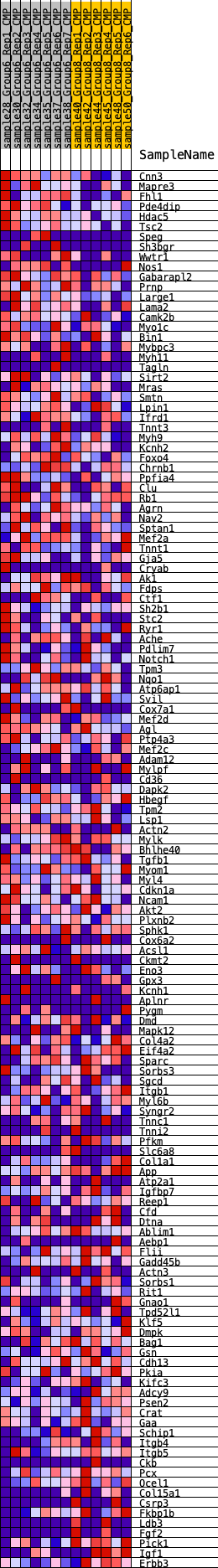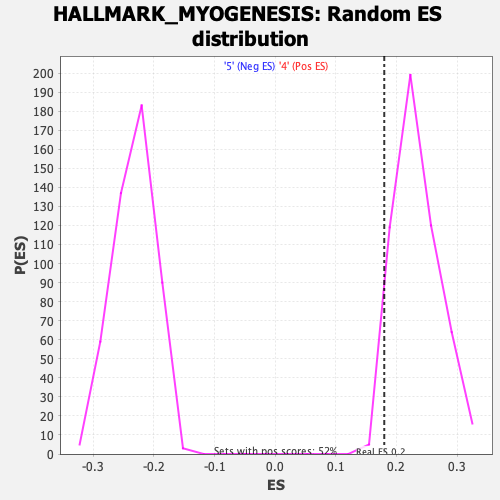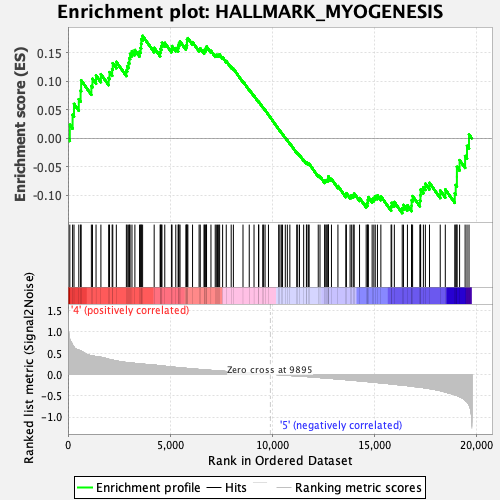
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group6_versus_Group8.CMP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | CMP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.17999148 |
| Normalized Enrichment Score (NES) | 0.768528 |
| Nominal p-value | 0.9617591 |
| FDR q-value | 0.89496976 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cnn3 | 89 | 0.809 | 0.0241 | Yes |
| 2 | Mapre3 | 225 | 0.689 | 0.0415 | Yes |
| 3 | Fhl1 | 294 | 0.638 | 0.0606 | Yes |
| 4 | Pde4dip | 529 | 0.570 | 0.0689 | Yes |
| 5 | Hdac5 | 617 | 0.555 | 0.0840 | Yes |
| 6 | Tsc2 | 643 | 0.545 | 0.1021 | Yes |
| 7 | Speg | 1144 | 0.439 | 0.0921 | Yes |
| 8 | Sh3bgr | 1198 | 0.433 | 0.1047 | Yes |
| 9 | Wwtr1 | 1373 | 0.418 | 0.1107 | Yes |
| 10 | Nos1 | 1611 | 0.401 | 0.1128 | Yes |
| 11 | Gabarapl2 | 1991 | 0.355 | 0.1060 | Yes |
| 12 | Prnp | 2033 | 0.350 | 0.1163 | Yes |
| 13 | Large1 | 2162 | 0.337 | 0.1217 | Yes |
| 14 | Lama2 | 2192 | 0.334 | 0.1320 | Yes |
| 15 | Camk2b | 2366 | 0.316 | 0.1344 | Yes |
| 16 | Myo1c | 2857 | 0.279 | 0.1193 | Yes |
| 17 | Bin1 | 2899 | 0.277 | 0.1270 | Yes |
| 18 | Mybpc3 | 2971 | 0.271 | 0.1330 | Yes |
| 19 | Myh11 | 3007 | 0.269 | 0.1407 | Yes |
| 20 | Tagln | 3040 | 0.268 | 0.1486 | Yes |
| 21 | Sirt2 | 3132 | 0.264 | 0.1533 | Yes |
| 22 | Mras | 3275 | 0.259 | 0.1552 | Yes |
| 23 | Smtn | 3503 | 0.252 | 0.1525 | Yes |
| 24 | Lpin1 | 3556 | 0.249 | 0.1587 | Yes |
| 25 | Ifrd1 | 3580 | 0.248 | 0.1663 | Yes |
| 26 | Tnnt3 | 3590 | 0.247 | 0.1745 | Yes |
| 27 | Myh9 | 3653 | 0.244 | 0.1800 | Yes |
| 28 | Kcnh2 | 4219 | 0.219 | 0.1589 | No |
| 29 | Foxo4 | 4511 | 0.202 | 0.1513 | No |
| 30 | Chrnb1 | 4531 | 0.200 | 0.1574 | No |
| 31 | Ppfia4 | 4572 | 0.198 | 0.1624 | No |
| 32 | Clu | 4601 | 0.197 | 0.1679 | No |
| 33 | Rb1 | 4738 | 0.192 | 0.1677 | No |
| 34 | Agrn | 5064 | 0.175 | 0.1574 | No |
| 35 | Nav2 | 5092 | 0.174 | 0.1621 | No |
| 36 | Sptan1 | 5277 | 0.165 | 0.1586 | No |
| 37 | Mef2a | 5395 | 0.158 | 0.1582 | No |
| 38 | Tnnt1 | 5403 | 0.158 | 0.1634 | No |
| 39 | Gja5 | 5438 | 0.156 | 0.1672 | No |
| 40 | Cryab | 5485 | 0.154 | 0.1703 | No |
| 41 | Ak1 | 5771 | 0.143 | 0.1609 | No |
| 42 | Fdps | 5807 | 0.142 | 0.1641 | No |
| 43 | Ctf1 | 5825 | 0.141 | 0.1682 | No |
| 44 | Sh2b1 | 5827 | 0.141 | 0.1732 | No |
| 45 | Stc2 | 5869 | 0.139 | 0.1760 | No |
| 46 | Ryr1 | 6098 | 0.129 | 0.1690 | No |
| 47 | Ache | 6419 | 0.117 | 0.1568 | No |
| 48 | Pdlim7 | 6475 | 0.115 | 0.1580 | No |
| 49 | Notch1 | 6663 | 0.108 | 0.1523 | No |
| 50 | Tpm3 | 6676 | 0.107 | 0.1555 | No |
| 51 | Nqo1 | 6740 | 0.105 | 0.1560 | No |
| 52 | Atp6ap1 | 6760 | 0.104 | 0.1587 | No |
| 53 | Svil | 6783 | 0.104 | 0.1613 | No |
| 54 | Cox7a1 | 6999 | 0.095 | 0.1537 | No |
| 55 | Mef2d | 7208 | 0.087 | 0.1462 | No |
| 56 | Agl | 7261 | 0.086 | 0.1466 | No |
| 57 | Ptp4a3 | 7322 | 0.083 | 0.1465 | No |
| 58 | Mef2c | 7376 | 0.082 | 0.1467 | No |
| 59 | Adam12 | 7424 | 0.080 | 0.1471 | No |
| 60 | Mylpf | 7570 | 0.075 | 0.1424 | No |
| 61 | Cd36 | 7748 | 0.070 | 0.1358 | No |
| 62 | Dapk2 | 7990 | 0.062 | 0.1257 | No |
| 63 | Hbegf | 8097 | 0.059 | 0.1224 | No |
| 64 | Tpm2 | 8568 | 0.044 | 0.1000 | No |
| 65 | Lsp1 | 8874 | 0.034 | 0.0857 | No |
| 66 | Actn2 | 9108 | 0.026 | 0.0748 | No |
| 67 | Mylk | 9328 | 0.018 | 0.0642 | No |
| 68 | Bhlhe40 | 9341 | 0.018 | 0.0643 | No |
| 69 | Tgfb1 | 9534 | 0.012 | 0.0549 | No |
| 70 | Myom1 | 9578 | 0.011 | 0.0531 | No |
| 71 | Myl4 | 9649 | 0.008 | 0.0499 | No |
| 72 | Cdkn1a | 9815 | 0.003 | 0.0415 | No |
| 73 | Ncam1 | 10314 | -0.003 | 0.0163 | No |
| 74 | Akt2 | 10373 | -0.005 | 0.0135 | No |
| 75 | Plxnb2 | 10445 | -0.007 | 0.0102 | No |
| 76 | Sphk1 | 10504 | -0.009 | 0.0075 | No |
| 77 | Cox6a2 | 10639 | -0.014 | 0.0012 | No |
| 78 | Acsl1 | 10739 | -0.016 | -0.0033 | No |
| 79 | Ckmt2 | 10862 | -0.021 | -0.0087 | No |
| 80 | Eno3 | 11198 | -0.032 | -0.0247 | No |
| 81 | Gpx3 | 11247 | -0.034 | -0.0259 | No |
| 82 | Kcnh1 | 11334 | -0.037 | -0.0290 | No |
| 83 | Aplnr | 11538 | -0.042 | -0.0379 | No |
| 84 | Pygm | 11673 | -0.046 | -0.0431 | No |
| 85 | Dmd | 11695 | -0.046 | -0.0425 | No |
| 86 | Mapk12 | 11768 | -0.049 | -0.0444 | No |
| 87 | Col4a2 | 11806 | -0.051 | -0.0445 | No |
| 88 | Eif4a2 | 12252 | -0.068 | -0.0648 | No |
| 89 | Sparc | 12338 | -0.071 | -0.0666 | No |
| 90 | Sorbs3 | 12567 | -0.079 | -0.0754 | No |
| 91 | Sgcd | 12585 | -0.080 | -0.0735 | No |
| 92 | Itgb1 | 12641 | -0.081 | -0.0734 | No |
| 93 | Myl6b | 12686 | -0.083 | -0.0727 | No |
| 94 | Syngr2 | 12740 | -0.085 | -0.0724 | No |
| 95 | Tnnc1 | 12744 | -0.085 | -0.0695 | No |
| 96 | Tnni2 | 12746 | -0.085 | -0.0666 | No |
| 97 | Pfkm | 12899 | -0.090 | -0.0711 | No |
| 98 | Slc6a8 | 13216 | -0.104 | -0.0835 | No |
| 99 | Col1a1 | 13602 | -0.119 | -0.0989 | No |
| 100 | App | 13631 | -0.120 | -0.0961 | No |
| 101 | Atp2a1 | 13812 | -0.128 | -0.1007 | No |
| 102 | Igfbp7 | 13889 | -0.131 | -0.1000 | No |
| 103 | Reep1 | 13966 | -0.135 | -0.0991 | No |
| 104 | Cfd | 14012 | -0.136 | -0.0966 | No |
| 105 | Dtna | 14274 | -0.148 | -0.1046 | No |
| 106 | Ablim1 | 14599 | -0.162 | -0.1154 | No |
| 107 | Aebp1 | 14660 | -0.164 | -0.1127 | No |
| 108 | Flii | 14683 | -0.165 | -0.1079 | No |
| 109 | Gadd45b | 14702 | -0.166 | -0.1030 | No |
| 110 | Actn3 | 14891 | -0.175 | -0.1063 | No |
| 111 | Sorbs1 | 14975 | -0.180 | -0.1042 | No |
| 112 | Rit1 | 15043 | -0.183 | -0.1012 | No |
| 113 | Gnao1 | 15151 | -0.187 | -0.1000 | No |
| 114 | Tpd52l1 | 15318 | -0.195 | -0.1015 | No |
| 115 | Klf5 | 15819 | -0.219 | -0.1193 | No |
| 116 | Dmpk | 15850 | -0.220 | -0.1130 | No |
| 117 | Bag1 | 15978 | -0.227 | -0.1115 | No |
| 118 | Gsn | 16364 | -0.245 | -0.1224 | No |
| 119 | Cdh13 | 16424 | -0.248 | -0.1167 | No |
| 120 | Pkia | 16619 | -0.259 | -0.1174 | No |
| 121 | Kifc3 | 16821 | -0.272 | -0.1180 | No |
| 122 | Adcy9 | 16825 | -0.272 | -0.1085 | No |
| 123 | Psen2 | 16872 | -0.275 | -0.1012 | No |
| 124 | Crat | 17229 | -0.295 | -0.1089 | No |
| 125 | Gaa | 17257 | -0.296 | -0.0998 | No |
| 126 | Schip1 | 17266 | -0.297 | -0.0897 | No |
| 127 | Itgb4 | 17400 | -0.307 | -0.0856 | No |
| 128 | Itgb5 | 17497 | -0.315 | -0.0794 | No |
| 129 | Ckb | 17699 | -0.330 | -0.0779 | No |
| 130 | Pcx | 18225 | -0.377 | -0.0913 | No |
| 131 | Ocel1 | 18473 | -0.409 | -0.0895 | No |
| 132 | Col15a1 | 18937 | -0.473 | -0.0963 | No |
| 133 | Csrp3 | 18983 | -0.480 | -0.0816 | No |
| 134 | Fkbp1b | 19041 | -0.492 | -0.0672 | No |
| 135 | Ldb3 | 19042 | -0.492 | -0.0498 | No |
| 136 | Fgf2 | 19173 | -0.517 | -0.0381 | No |
| 137 | Pick1 | 19443 | -0.594 | -0.0308 | No |
| 138 | Igf1 | 19536 | -0.636 | -0.0130 | No |
| 139 | Erbb3 | 19634 | -0.705 | 0.0070 | No |