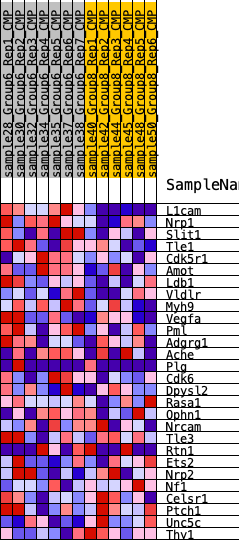
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group6_versus_Group8.CMP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | CMP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
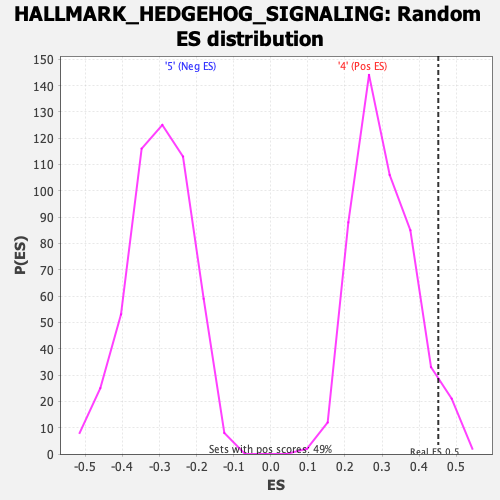
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.45174515 |
| Normalized Enrichment Score (NES) | 1.4762305 |
| Nominal p-value | 0.05882353 |
| FDR q-value | 0.46775138 |
| FWER p-Value | 0.399 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | L1cam | 3 | 1.359 | 0.1836 | Yes |
| 2 | Nrp1 | 535 | 0.568 | 0.2335 | Yes |
| 3 | Slit1 | 904 | 0.476 | 0.2792 | Yes |
| 4 | Tle1 | 1081 | 0.446 | 0.3306 | Yes |
| 5 | Cdk5r1 | 1304 | 0.423 | 0.3766 | Yes |
| 6 | Amot | 1662 | 0.400 | 0.4125 | Yes |
| 7 | Ldb1 | 2475 | 0.306 | 0.4128 | Yes |
| 8 | Vldlr | 3093 | 0.267 | 0.4177 | Yes |
| 9 | Myh9 | 3653 | 0.244 | 0.4224 | Yes |
| 10 | Vegfa | 3713 | 0.239 | 0.4517 | Yes |
| 11 | Pml | 5921 | 0.137 | 0.3585 | No |
| 12 | Adgrg1 | 6005 | 0.133 | 0.3723 | No |
| 13 | Ache | 6419 | 0.117 | 0.3672 | No |
| 14 | Plg | 6595 | 0.111 | 0.3733 | No |
| 15 | Cdk6 | 6652 | 0.108 | 0.3851 | No |
| 16 | Dpysl2 | 7178 | 0.089 | 0.3705 | No |
| 17 | Rasa1 | 7584 | 0.075 | 0.3600 | No |
| 18 | Ophn1 | 8795 | 0.036 | 0.3036 | No |
| 19 | Nrcam | 11279 | -0.035 | 0.1826 | No |
| 20 | Tle3 | 11304 | -0.036 | 0.1862 | No |
| 21 | Rtn1 | 11614 | -0.044 | 0.1765 | No |
| 22 | Ets2 | 12586 | -0.080 | 0.1382 | No |
| 23 | Nrp2 | 12997 | -0.095 | 0.1302 | No |
| 24 | Nf1 | 14584 | -0.161 | 0.0716 | No |
| 25 | Celsr1 | 14658 | -0.164 | 0.0902 | No |
| 26 | Ptch1 | 15745 | -0.215 | 0.0642 | No |
| 27 | Unc5c | 19062 | -0.494 | -0.0369 | No |
| 28 | Thy1 | 19264 | -0.538 | 0.0257 | No |