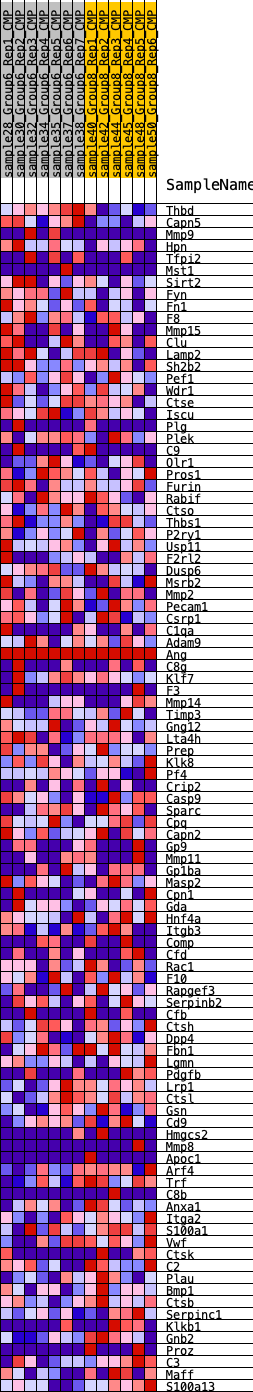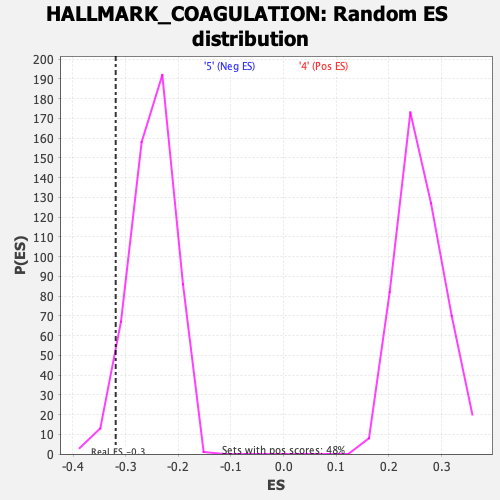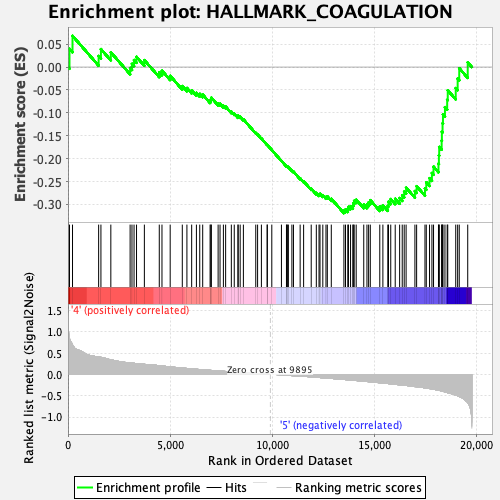
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group6_versus_Group8.CMP_Pheno.cls#Group6_versus_Group8_repos |
| Phenotype | CMP_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.3189236 |
| Normalized Enrichment Score (NES) | -1.2803055 |
| Nominal p-value | 0.03269231 |
| FDR q-value | 0.28013217 |
| FWER p-Value | 0.788 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Thbd | 73 | 0.846 | 0.0398 | No |
| 2 | Capn5 | 221 | 0.695 | 0.0681 | No |
| 3 | Mmp9 | 1500 | 0.409 | 0.0241 | No |
| 4 | Hpn | 1612 | 0.401 | 0.0391 | No |
| 5 | Tfpi2 | 2097 | 0.343 | 0.0322 | No |
| 6 | Mst1 | 3041 | 0.268 | -0.0020 | No |
| 7 | Sirt2 | 3132 | 0.264 | 0.0070 | No |
| 8 | Fyn | 3234 | 0.262 | 0.0153 | No |
| 9 | Fn1 | 3355 | 0.254 | 0.0223 | No |
| 10 | F8 | 3739 | 0.238 | 0.0151 | No |
| 11 | Mmp15 | 4469 | 0.205 | -0.0115 | No |
| 12 | Clu | 4601 | 0.197 | -0.0080 | No |
| 13 | Lamp2 | 5003 | 0.178 | -0.0192 | No |
| 14 | Sh2b2 | 5596 | 0.151 | -0.0416 | No |
| 15 | Pef1 | 5822 | 0.141 | -0.0457 | No |
| 16 | Wdr1 | 6055 | 0.131 | -0.0508 | No |
| 17 | Ctse | 6287 | 0.122 | -0.0563 | No |
| 18 | Iscu | 6450 | 0.116 | -0.0586 | No |
| 19 | Plg | 6595 | 0.111 | -0.0602 | No |
| 20 | Plek | 6957 | 0.097 | -0.0736 | No |
| 21 | C9 | 6994 | 0.095 | -0.0705 | No |
| 22 | Olr1 | 7018 | 0.095 | -0.0668 | No |
| 23 | Pros1 | 7350 | 0.083 | -0.0794 | No |
| 24 | Furin | 7444 | 0.080 | -0.0800 | No |
| 25 | Rabif | 7608 | 0.074 | -0.0845 | No |
| 26 | Ctso | 7718 | 0.070 | -0.0864 | No |
| 27 | Thbs1 | 7997 | 0.062 | -0.0974 | No |
| 28 | P2ry1 | 8140 | 0.057 | -0.1016 | No |
| 29 | Usp11 | 8324 | 0.051 | -0.1083 | No |
| 30 | F2rl2 | 8328 | 0.051 | -0.1058 | No |
| 31 | Dusp6 | 8430 | 0.048 | -0.1085 | No |
| 32 | Msrb2 | 8587 | 0.044 | -0.1142 | No |
| 33 | Mmp2 | 9193 | 0.023 | -0.1438 | No |
| 34 | Pecam1 | 9283 | 0.020 | -0.1472 | No |
| 35 | Csrp1 | 9469 | 0.014 | -0.1559 | No |
| 36 | C1qa | 9754 | 0.005 | -0.1701 | No |
| 37 | Adam9 | 9755 | 0.005 | -0.1699 | No |
| 38 | Ang | 9978 | 0.000 | -0.1811 | No |
| 39 | C8g | 10450 | -0.007 | -0.2047 | No |
| 40 | Klf7 | 10696 | -0.015 | -0.2164 | No |
| 41 | F3 | 10714 | -0.016 | -0.2164 | No |
| 42 | Mmp14 | 10754 | -0.017 | -0.2176 | No |
| 43 | Timp3 | 10789 | -0.018 | -0.2183 | No |
| 44 | Gng12 | 10962 | -0.025 | -0.2258 | No |
| 45 | Lta4h | 11039 | -0.027 | -0.2283 | No |
| 46 | Prep | 11366 | -0.037 | -0.2429 | No |
| 47 | Klk8 | 11536 | -0.042 | -0.2494 | No |
| 48 | Pf4 | 11908 | -0.055 | -0.2654 | No |
| 49 | Crip2 | 12158 | -0.064 | -0.2748 | No |
| 50 | Casp9 | 12290 | -0.069 | -0.2779 | No |
| 51 | Sparc | 12338 | -0.071 | -0.2766 | No |
| 52 | Cpq | 12476 | -0.075 | -0.2797 | No |
| 53 | Capn2 | 12631 | -0.081 | -0.2834 | No |
| 54 | Gp9 | 12697 | -0.084 | -0.2824 | No |
| 55 | Mmp11 | 12891 | -0.090 | -0.2876 | No |
| 56 | Gp1ba | 13509 | -0.116 | -0.3130 | Yes |
| 57 | Masp2 | 13586 | -0.118 | -0.3108 | Yes |
| 58 | Cpn1 | 13700 | -0.123 | -0.3102 | Yes |
| 59 | Gda | 13731 | -0.125 | -0.3053 | Yes |
| 60 | Hnf4a | 13834 | -0.129 | -0.3038 | Yes |
| 61 | Itgb3 | 13947 | -0.134 | -0.3026 | Yes |
| 62 | Comp | 13970 | -0.135 | -0.2968 | Yes |
| 63 | Cfd | 14012 | -0.136 | -0.2919 | Yes |
| 64 | Rac1 | 14110 | -0.141 | -0.2896 | Yes |
| 65 | F10 | 14480 | -0.156 | -0.3003 | Yes |
| 66 | Rapgef3 | 14637 | -0.163 | -0.2999 | Yes |
| 67 | Serpinb2 | 14719 | -0.167 | -0.2954 | Yes |
| 68 | Cfb | 14801 | -0.171 | -0.2907 | Yes |
| 69 | Ctsh | 15265 | -0.192 | -0.3043 | Yes |
| 70 | Dpp4 | 15417 | -0.199 | -0.3018 | Yes |
| 71 | Fbn1 | 15658 | -0.211 | -0.3031 | Yes |
| 72 | Lgmn | 15692 | -0.212 | -0.2939 | Yes |
| 73 | Pdgfb | 15802 | -0.218 | -0.2882 | Yes |
| 74 | Lrp1 | 16021 | -0.230 | -0.2875 | Yes |
| 75 | Ctsl | 16237 | -0.242 | -0.2860 | Yes |
| 76 | Gsn | 16364 | -0.245 | -0.2798 | Yes |
| 77 | Cd9 | 16461 | -0.250 | -0.2718 | Yes |
| 78 | Hmgcs2 | 16554 | -0.256 | -0.2633 | Yes |
| 79 | Mmp8 | 16985 | -0.283 | -0.2706 | Yes |
| 80 | Apoc1 | 17069 | -0.285 | -0.2602 | Yes |
| 81 | Arf4 | 17478 | -0.314 | -0.2648 | Yes |
| 82 | Trf | 17542 | -0.318 | -0.2516 | Yes |
| 83 | C8b | 17701 | -0.330 | -0.2427 | Yes |
| 84 | Anxa1 | 17818 | -0.337 | -0.2312 | Yes |
| 85 | Itga2 | 17897 | -0.345 | -0.2175 | Yes |
| 86 | S100a1 | 18139 | -0.367 | -0.2108 | Yes |
| 87 | Vwf | 18160 | -0.370 | -0.1928 | Yes |
| 88 | Ctsk | 18175 | -0.371 | -0.1744 | Yes |
| 89 | C2 | 18294 | -0.385 | -0.1606 | Yes |
| 90 | Plau | 18306 | -0.386 | -0.1414 | Yes |
| 91 | Bmp1 | 18339 | -0.391 | -0.1229 | Yes |
| 92 | Ctsb | 18359 | -0.394 | -0.1036 | Yes |
| 93 | Serpinc1 | 18456 | -0.406 | -0.0876 | Yes |
| 94 | Klkb1 | 18565 | -0.422 | -0.0714 | Yes |
| 95 | Gnb2 | 18593 | -0.426 | -0.0508 | Yes |
| 96 | Proz | 18984 | -0.480 | -0.0459 | Yes |
| 97 | C3 | 19078 | -0.497 | -0.0251 | Yes |
| 98 | Maff | 19156 | -0.513 | -0.0027 | Yes |
| 99 | S100a13 | 19571 | -0.659 | 0.0102 | Yes |