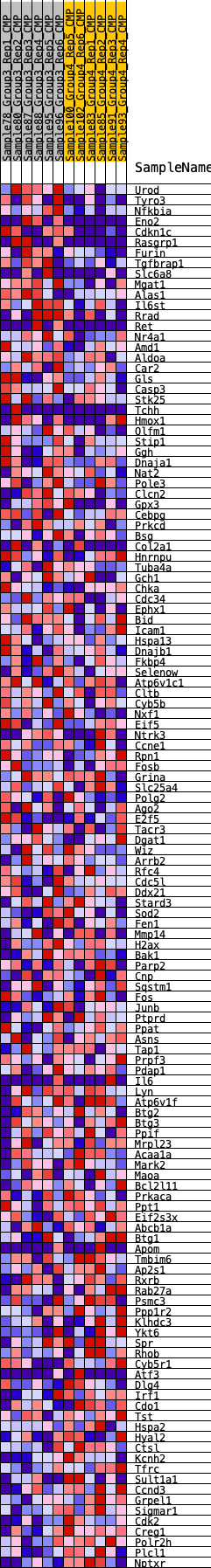
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group3_versus_Group4.CMP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
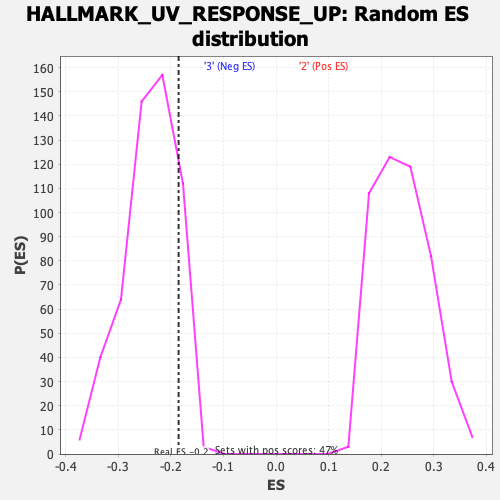
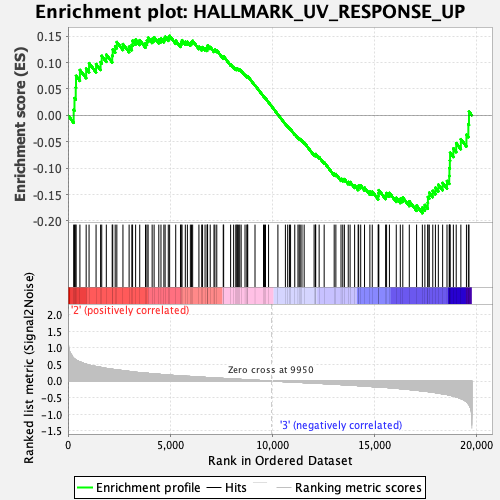
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | -0.18528718 |
| Normalized Enrichment Score (NES) | -0.7789834 |
| Nominal p-value | 0.8560606 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Urod | 278 | 0.685 | 0.0106 | No |
| 2 | Tyro3 | 317 | 0.664 | 0.0326 | No |
| 3 | Nfkbia | 379 | 0.639 | 0.0526 | No |
| 4 | Eno2 | 393 | 0.634 | 0.0748 | No |
| 5 | Cdkn1c | 583 | 0.575 | 0.0859 | No |
| 6 | Rasgrp1 | 889 | 0.507 | 0.0887 | No |
| 7 | Furin | 1031 | 0.480 | 0.0988 | No |
| 8 | Tgfbrap1 | 1374 | 0.435 | 0.0971 | No |
| 9 | Slc6a8 | 1596 | 0.413 | 0.1008 | No |
| 10 | Mgat1 | 1652 | 0.405 | 0.1126 | No |
| 11 | Alas1 | 1876 | 0.380 | 0.1150 | No |
| 12 | Il6st | 2164 | 0.355 | 0.1132 | No |
| 13 | Rrad | 2190 | 0.352 | 0.1246 | No |
| 14 | Ret | 2310 | 0.340 | 0.1308 | No |
| 15 | Nr4a1 | 2391 | 0.334 | 0.1388 | No |
| 16 | Amd1 | 2690 | 0.312 | 0.1349 | No |
| 17 | Aldoa | 2992 | 0.290 | 0.1301 | No |
| 18 | Car2 | 3126 | 0.277 | 0.1333 | No |
| 19 | Gls | 3162 | 0.274 | 0.1414 | No |
| 20 | Casp3 | 3311 | 0.264 | 0.1434 | No |
| 21 | Stk25 | 3512 | 0.254 | 0.1423 | No |
| 22 | Tchh | 3794 | 0.237 | 0.1366 | No |
| 23 | Hmox1 | 3864 | 0.234 | 0.1416 | No |
| 24 | Olfm1 | 3922 | 0.232 | 0.1470 | No |
| 25 | Stip1 | 4120 | 0.220 | 0.1449 | No |
| 26 | Ggh | 4216 | 0.215 | 0.1479 | No |
| 27 | Dnaja1 | 4446 | 0.204 | 0.1436 | No |
| 28 | Nat2 | 4552 | 0.198 | 0.1454 | No |
| 29 | Pole3 | 4694 | 0.191 | 0.1451 | No |
| 30 | Clcn2 | 4755 | 0.188 | 0.1488 | No |
| 31 | Gpx3 | 4910 | 0.180 | 0.1475 | No |
| 32 | Cebpg | 4977 | 0.177 | 0.1505 | No |
| 33 | Prkcd | 5274 | 0.165 | 0.1414 | No |
| 34 | Bsg | 5507 | 0.155 | 0.1351 | No |
| 35 | Col2a1 | 5544 | 0.152 | 0.1388 | No |
| 36 | Hnrnpu | 5583 | 0.150 | 0.1423 | No |
| 37 | Tuba4a | 5744 | 0.144 | 0.1394 | No |
| 38 | Gch1 | 5845 | 0.140 | 0.1393 | No |
| 39 | Chka | 5991 | 0.138 | 0.1369 | No |
| 40 | Cdc34 | 6048 | 0.135 | 0.1390 | No |
| 41 | Ephx1 | 6103 | 0.134 | 0.1410 | No |
| 42 | Bid | 6410 | 0.122 | 0.1298 | No |
| 43 | Icam1 | 6547 | 0.116 | 0.1271 | No |
| 44 | Hspa13 | 6588 | 0.115 | 0.1292 | No |
| 45 | Dnajb1 | 6710 | 0.109 | 0.1270 | No |
| 46 | Fkbp4 | 6810 | 0.106 | 0.1258 | No |
| 47 | Selenow | 6824 | 0.105 | 0.1289 | No |
| 48 | Atp6v1c1 | 6835 | 0.105 | 0.1322 | No |
| 49 | Cltb | 6963 | 0.101 | 0.1294 | No |
| 50 | Cyb5b | 7148 | 0.094 | 0.1234 | No |
| 51 | Nxf1 | 7193 | 0.092 | 0.1245 | No |
| 52 | Eif5 | 7284 | 0.089 | 0.1231 | No |
| 53 | Ntrk3 | 7601 | 0.078 | 0.1098 | No |
| 54 | Ccne1 | 7621 | 0.078 | 0.1117 | No |
| 55 | Rpn1 | 7962 | 0.066 | 0.0967 | No |
| 56 | Fosb | 8110 | 0.061 | 0.0915 | No |
| 57 | Grina | 8215 | 0.058 | 0.0882 | No |
| 58 | Slc25a4 | 8264 | 0.056 | 0.0878 | No |
| 59 | Polg2 | 8292 | 0.055 | 0.0884 | No |
| 60 | Ago2 | 8355 | 0.053 | 0.0872 | No |
| 61 | E2f5 | 8408 | 0.051 | 0.0864 | No |
| 62 | Tacr3 | 8477 | 0.048 | 0.0847 | No |
| 63 | Dgat1 | 8667 | 0.042 | 0.0766 | No |
| 64 | Wiz | 8757 | 0.039 | 0.0735 | No |
| 65 | Arrb2 | 8780 | 0.039 | 0.0737 | No |
| 66 | Rfc4 | 8800 | 0.038 | 0.0742 | No |
| 67 | Cdc5l | 9156 | 0.025 | 0.0570 | No |
| 68 | Ddx21 | 9572 | 0.012 | 0.0363 | No |
| 69 | Stard3 | 9622 | 0.011 | 0.0342 | No |
| 70 | Sod2 | 9670 | 0.009 | 0.0321 | No |
| 71 | Fen1 | 9820 | 0.004 | 0.0247 | No |
| 72 | Mmp14 | 10277 | -0.006 | 0.0017 | No |
| 73 | H2ax | 10644 | -0.018 | -0.0163 | No |
| 74 | Bak1 | 10756 | -0.021 | -0.0212 | No |
| 75 | Parp2 | 10842 | -0.024 | -0.0247 | No |
| 76 | Cnp | 10897 | -0.025 | -0.0265 | No |
| 77 | Sqstm1 | 11102 | -0.032 | -0.0357 | No |
| 78 | Fos | 11260 | -0.037 | -0.0424 | No |
| 79 | Junb | 11318 | -0.039 | -0.0439 | No |
| 80 | Ptprd | 11369 | -0.040 | -0.0450 | No |
| 81 | Ppat | 11454 | -0.042 | -0.0478 | No |
| 82 | Asns | 11563 | -0.046 | -0.0516 | No |
| 83 | Tap1 | 12050 | -0.062 | -0.0741 | No |
| 84 | Prpf3 | 12116 | -0.064 | -0.0751 | No |
| 85 | Pdap1 | 12128 | -0.065 | -0.0733 | No |
| 86 | Il6 | 12296 | -0.070 | -0.0793 | No |
| 87 | Lyn | 12542 | -0.076 | -0.0890 | No |
| 88 | Atp6v1f | 13033 | -0.092 | -0.1107 | No |
| 89 | Btg2 | 13115 | -0.095 | -0.1114 | No |
| 90 | Btg3 | 13367 | -0.104 | -0.1204 | No |
| 91 | Ppif | 13453 | -0.107 | -0.1208 | No |
| 92 | Mrpl23 | 13532 | -0.110 | -0.1208 | No |
| 93 | Acaa1a | 13721 | -0.118 | -0.1261 | No |
| 94 | Mark2 | 13812 | -0.121 | -0.1264 | No |
| 95 | Maoa | 14033 | -0.129 | -0.1329 | No |
| 96 | Bcl2l11 | 14206 | -0.137 | -0.1367 | No |
| 97 | Prkaca | 14231 | -0.137 | -0.1330 | No |
| 98 | Ppt1 | 14331 | -0.141 | -0.1329 | No |
| 99 | Eif2s3x | 14518 | -0.148 | -0.1371 | No |
| 100 | Abcb1a | 14780 | -0.160 | -0.1446 | No |
| 101 | Btg1 | 14897 | -0.166 | -0.1445 | No |
| 102 | Apom | 15186 | -0.175 | -0.1529 | No |
| 103 | Tmbim6 | 15202 | -0.175 | -0.1473 | No |
| 104 | Ap2s1 | 15218 | -0.176 | -0.1417 | No |
| 105 | Rxrb | 15551 | -0.190 | -0.1518 | No |
| 106 | Rab27a | 15598 | -0.192 | -0.1472 | No |
| 107 | Psmc3 | 15737 | -0.199 | -0.1470 | No |
| 108 | Ppp1r2 | 16072 | -0.217 | -0.1562 | No |
| 109 | Klhdc3 | 16270 | -0.227 | -0.1580 | No |
| 110 | Ykt6 | 16392 | -0.233 | -0.1558 | No |
| 111 | Spr | 16710 | -0.253 | -0.1628 | No |
| 112 | Rhob | 17067 | -0.276 | -0.1709 | No |
| 113 | Cyb5r1 | 17350 | -0.297 | -0.1746 | Yes |
| 114 | Atf3 | 17473 | -0.305 | -0.1698 | Yes |
| 115 | Dlg4 | 17616 | -0.314 | -0.1657 | Yes |
| 116 | Irf1 | 17623 | -0.315 | -0.1546 | Yes |
| 117 | Cdo1 | 17689 | -0.319 | -0.1464 | Yes |
| 118 | Tst | 17860 | -0.333 | -0.1430 | Yes |
| 119 | Hspa2 | 17989 | -0.347 | -0.1370 | Yes |
| 120 | Hyal2 | 18132 | -0.360 | -0.1313 | Yes |
| 121 | Ctsl | 18343 | -0.384 | -0.1281 | Yes |
| 122 | Kcnh2 | 18556 | -0.403 | -0.1244 | Yes |
| 123 | Tfrc | 18666 | -0.420 | -0.1147 | Yes |
| 124 | Sult1a1 | 18675 | -0.421 | -0.0999 | Yes |
| 125 | Ccnd3 | 18696 | -0.424 | -0.0857 | Yes |
| 126 | Grpel1 | 18707 | -0.425 | -0.0708 | Yes |
| 127 | Sigmar1 | 18868 | -0.450 | -0.0628 | Yes |
| 128 | Cdk2 | 19007 | -0.475 | -0.0526 | Yes |
| 129 | Creg1 | 19235 | -0.527 | -0.0452 | Yes |
| 130 | Polr2h | 19508 | -0.619 | -0.0367 | Yes |
| 131 | Plcl1 | 19602 | -0.681 | -0.0168 | Yes |
| 132 | Nptxr | 19633 | -0.704 | 0.0070 | Yes |