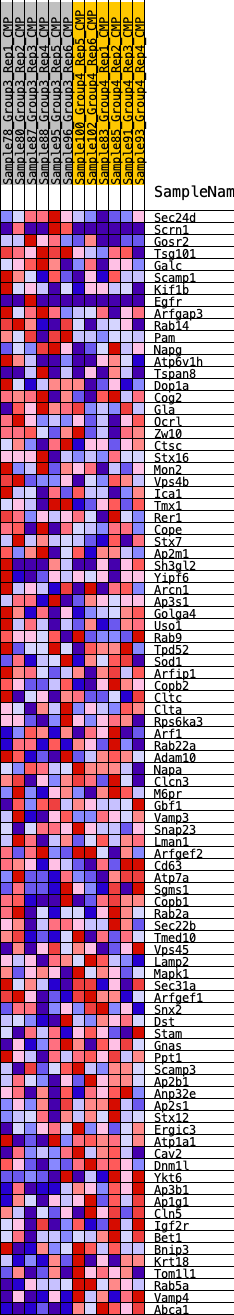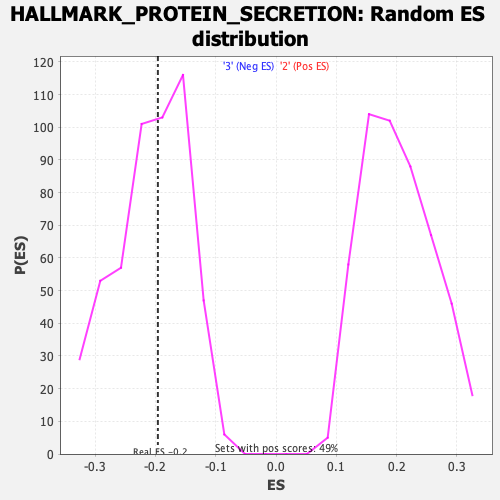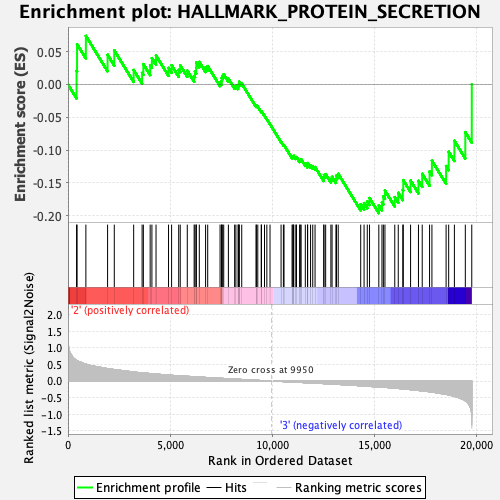
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group3_versus_Group4.CMP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.19597298 |
| Normalized Enrichment Score (NES) | -0.9479401 |
| Nominal p-value | 0.53125 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Sec24d | 419 | 0.623 | 0.0209 | No |
| 2 | Scrn1 | 444 | 0.615 | 0.0613 | No |
| 3 | Gosr2 | 875 | 0.510 | 0.0740 | No |
| 4 | Tsg101 | 1938 | 0.373 | 0.0453 | No |
| 5 | Galc | 2266 | 0.344 | 0.0519 | No |
| 6 | Scamp1 | 3215 | 0.270 | 0.0221 | No |
| 7 | Kif1b | 3628 | 0.245 | 0.0177 | No |
| 8 | Egfr | 3690 | 0.242 | 0.0310 | No |
| 9 | Arfgap3 | 4023 | 0.226 | 0.0294 | No |
| 10 | Rab14 | 4108 | 0.221 | 0.0401 | No |
| 11 | Pam | 4310 | 0.210 | 0.0441 | No |
| 12 | Napg | 4922 | 0.180 | 0.0252 | No |
| 13 | Atp6v1h | 5072 | 0.172 | 0.0293 | No |
| 14 | Tspan8 | 5420 | 0.158 | 0.0224 | No |
| 15 | Dop1a | 5495 | 0.155 | 0.0291 | No |
| 16 | Cog2 | 5840 | 0.140 | 0.0211 | No |
| 17 | Gla | 6176 | 0.131 | 0.0129 | No |
| 18 | Ocrl | 6217 | 0.129 | 0.0196 | No |
| 19 | Zw10 | 6277 | 0.127 | 0.0252 | No |
| 20 | Ctsc | 6282 | 0.127 | 0.0336 | No |
| 21 | Stx16 | 6429 | 0.121 | 0.0344 | No |
| 22 | Mon2 | 6736 | 0.108 | 0.0262 | No |
| 23 | Vps4b | 6840 | 0.105 | 0.0280 | No |
| 24 | Ica1 | 7445 | 0.084 | 0.0030 | No |
| 25 | Tmx1 | 7514 | 0.081 | 0.0050 | No |
| 26 | Rer1 | 7527 | 0.081 | 0.0098 | No |
| 27 | Cope | 7573 | 0.079 | 0.0129 | No |
| 28 | Stx7 | 7624 | 0.077 | 0.0156 | No |
| 29 | Ap2m1 | 7853 | 0.070 | 0.0088 | No |
| 30 | Sh3gl2 | 8158 | 0.060 | -0.0026 | No |
| 31 | Yipf6 | 8220 | 0.057 | -0.0018 | No |
| 32 | Arcn1 | 8326 | 0.054 | -0.0035 | No |
| 33 | Ap3s1 | 8351 | 0.053 | -0.0012 | No |
| 34 | Golga4 | 8376 | 0.052 | 0.0011 | No |
| 35 | Uso1 | 8382 | 0.052 | 0.0044 | No |
| 36 | Rab9 | 8506 | 0.047 | 0.0014 | No |
| 37 | Tpd52 | 9208 | 0.024 | -0.0327 | No |
| 38 | Sod1 | 9223 | 0.023 | -0.0318 | No |
| 39 | Arfip1 | 9292 | 0.021 | -0.0338 | No |
| 40 | Copb2 | 9460 | 0.016 | -0.0412 | No |
| 41 | Cltc | 9474 | 0.016 | -0.0408 | No |
| 42 | Clta | 9621 | 0.011 | -0.0475 | No |
| 43 | Rps6ka3 | 9736 | 0.007 | -0.0528 | No |
| 44 | Arf1 | 9891 | 0.002 | -0.0605 | No |
| 45 | Rab22a | 10432 | -0.011 | -0.0872 | No |
| 46 | Adam10 | 10546 | -0.015 | -0.0920 | No |
| 47 | Napa | 10577 | -0.016 | -0.0924 | No |
| 48 | Clcn3 | 10971 | -0.027 | -0.1105 | No |
| 49 | M6pr | 11007 | -0.029 | -0.1103 | No |
| 50 | Gbf1 | 11037 | -0.030 | -0.1098 | No |
| 51 | Vamp3 | 11055 | -0.031 | -0.1086 | No |
| 52 | Snap23 | 11155 | -0.034 | -0.1113 | No |
| 53 | Lman1 | 11187 | -0.035 | -0.1106 | No |
| 54 | Arfgef2 | 11340 | -0.039 | -0.1156 | No |
| 55 | Cd63 | 11372 | -0.040 | -0.1145 | No |
| 56 | Atp7a | 11414 | -0.041 | -0.1138 | No |
| 57 | Sgms1 | 11619 | -0.048 | -0.1209 | No |
| 58 | Copb1 | 11727 | -0.052 | -0.1228 | No |
| 59 | Rab2a | 11734 | -0.052 | -0.1196 | No |
| 60 | Sec22b | 11876 | -0.056 | -0.1230 | No |
| 61 | Tmed10 | 11983 | -0.060 | -0.1243 | No |
| 62 | Vps45 | 12096 | -0.063 | -0.1257 | No |
| 63 | Lamp2 | 12519 | -0.075 | -0.1421 | No |
| 64 | Mapk1 | 12535 | -0.075 | -0.1377 | No |
| 65 | Sec31a | 12615 | -0.078 | -0.1365 | No |
| 66 | Arfgef1 | 12871 | -0.086 | -0.1436 | No |
| 67 | Snx2 | 12929 | -0.089 | -0.1405 | No |
| 68 | Dst | 13119 | -0.095 | -0.1437 | No |
| 69 | Stam | 13146 | -0.096 | -0.1385 | No |
| 70 | Gnas | 13230 | -0.099 | -0.1360 | No |
| 71 | Ppt1 | 14331 | -0.141 | -0.1823 | No |
| 72 | Scamp3 | 14496 | -0.148 | -0.1807 | No |
| 73 | Ap2b1 | 14653 | -0.154 | -0.1781 | No |
| 74 | Anp32e | 14763 | -0.159 | -0.1729 | No |
| 75 | Ap2s1 | 15218 | -0.176 | -0.1841 | Yes |
| 76 | Stx12 | 15372 | -0.182 | -0.1795 | Yes |
| 77 | Ergic3 | 15441 | -0.186 | -0.1704 | Yes |
| 78 | Atp1a1 | 15518 | -0.189 | -0.1615 | Yes |
| 79 | Cav2 | 15999 | -0.213 | -0.1715 | Yes |
| 80 | Dnm1l | 16170 | -0.223 | -0.1650 | Yes |
| 81 | Ykt6 | 16392 | -0.233 | -0.1604 | Yes |
| 82 | Ap3b1 | 16416 | -0.235 | -0.1457 | Yes |
| 83 | Ap1g1 | 16773 | -0.258 | -0.1463 | Yes |
| 84 | Cln5 | 17159 | -0.282 | -0.1468 | Yes |
| 85 | Igf2r | 17343 | -0.296 | -0.1360 | Yes |
| 86 | Bet1 | 17702 | -0.320 | -0.1325 | Yes |
| 87 | Bnip3 | 17818 | -0.330 | -0.1160 | Yes |
| 88 | Krt18 | 18512 | -0.399 | -0.1243 | Yes |
| 89 | Tom1l1 | 18638 | -0.416 | -0.1024 | Yes |
| 90 | Rab5a | 18918 | -0.457 | -0.0857 | Yes |
| 91 | Vamp4 | 19453 | -0.595 | -0.0726 | Yes |
| 92 | Abca1 | 19769 | -1.310 | 0.0001 | Yes |