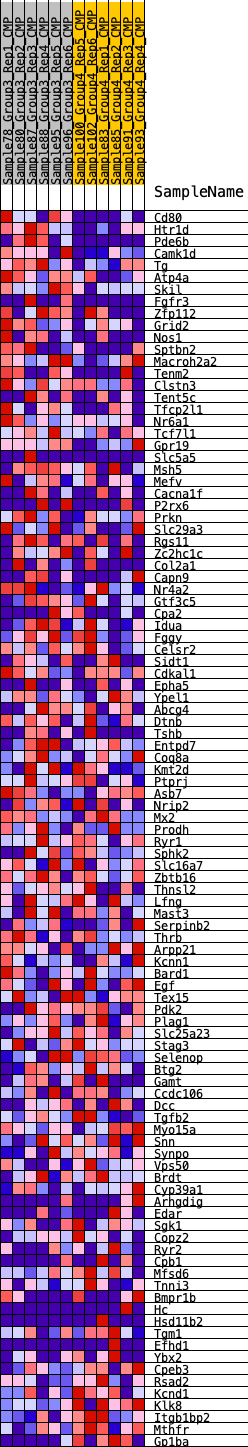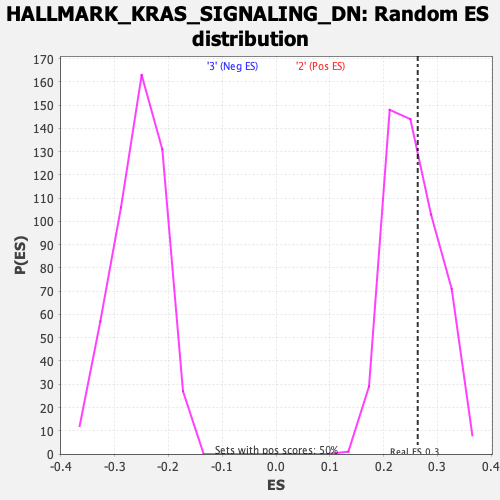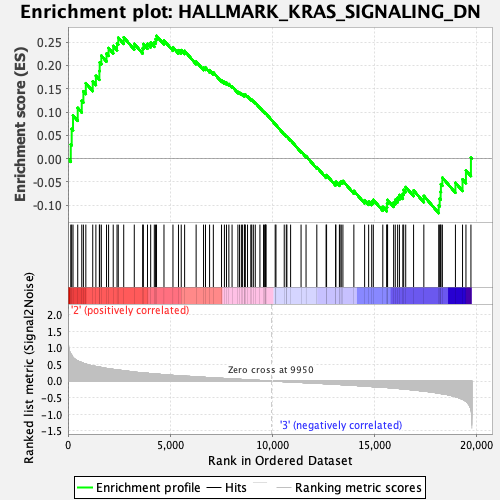
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group3_versus_Group4.CMP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.26347312 |
| Normalized Enrichment Score (NES) | 1.0378102 |
| Nominal p-value | 0.38690478 |
| FDR q-value | 0.92381865 |
| FWER p-Value | 0.968 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cd80 | 136 | 0.816 | 0.0304 | Yes |
| 2 | Htr1d | 181 | 0.778 | 0.0638 | Yes |
| 3 | Pde6b | 246 | 0.705 | 0.0928 | Yes |
| 4 | Camk1d | 476 | 0.606 | 0.1089 | Yes |
| 5 | Tg | 674 | 0.553 | 0.1242 | Yes |
| 6 | Atp4a | 754 | 0.533 | 0.1446 | Yes |
| 7 | Skil | 873 | 0.510 | 0.1619 | Yes |
| 8 | Fgfr3 | 1211 | 0.454 | 0.1656 | Yes |
| 9 | Zfp112 | 1365 | 0.436 | 0.1777 | Yes |
| 10 | Grid2 | 1536 | 0.418 | 0.1882 | Yes |
| 11 | Nos1 | 1552 | 0.416 | 0.2065 | Yes |
| 12 | Sptbn2 | 1632 | 0.408 | 0.2211 | Yes |
| 13 | Macroh2a2 | 1886 | 0.379 | 0.2256 | Yes |
| 14 | Tenm2 | 1987 | 0.369 | 0.2374 | Yes |
| 15 | Clstn3 | 2214 | 0.350 | 0.2419 | Yes |
| 16 | Tent5c | 2401 | 0.333 | 0.2477 | Yes |
| 17 | Tfcp2l1 | 2461 | 0.329 | 0.2597 | Yes |
| 18 | Nr6a1 | 2728 | 0.309 | 0.2603 | Yes |
| 19 | Tcf7l1 | 3245 | 0.268 | 0.2464 | Yes |
| 20 | Gpr19 | 3655 | 0.244 | 0.2367 | Yes |
| 21 | Slc5a5 | 3689 | 0.242 | 0.2461 | Yes |
| 22 | Msh5 | 3892 | 0.233 | 0.2465 | Yes |
| 23 | Mefv | 4048 | 0.224 | 0.2489 | Yes |
| 24 | Cacna1f | 4226 | 0.214 | 0.2497 | Yes |
| 25 | P2rx6 | 4287 | 0.211 | 0.2563 | Yes |
| 26 | Prkn | 4334 | 0.208 | 0.2635 | Yes |
| 27 | Slc29a3 | 4699 | 0.191 | 0.2537 | No |
| 28 | Rgs11 | 5139 | 0.169 | 0.2391 | No |
| 29 | Zc2hc1c | 5409 | 0.159 | 0.2327 | No |
| 30 | Col2a1 | 5544 | 0.152 | 0.2329 | No |
| 31 | Capn9 | 5710 | 0.146 | 0.2312 | No |
| 32 | Nr4a2 | 6273 | 0.127 | 0.2084 | No |
| 33 | Gtf3c5 | 6636 | 0.113 | 0.1952 | No |
| 34 | Cpa2 | 6732 | 0.109 | 0.1953 | No |
| 35 | Idua | 6932 | 0.102 | 0.1899 | No |
| 36 | Fggy | 7116 | 0.095 | 0.1849 | No |
| 37 | Celsr2 | 7517 | 0.081 | 0.1683 | No |
| 38 | Sidt1 | 7658 | 0.076 | 0.1646 | No |
| 39 | Cdkal1 | 7759 | 0.072 | 0.1628 | No |
| 40 | Epha5 | 7876 | 0.069 | 0.1601 | No |
| 41 | Ypel1 | 8033 | 0.064 | 0.1551 | No |
| 42 | Abcg4 | 8318 | 0.054 | 0.1431 | No |
| 43 | Dtnb | 8399 | 0.052 | 0.1414 | No |
| 44 | Tshb | 8489 | 0.048 | 0.1391 | No |
| 45 | Entpd7 | 8549 | 0.046 | 0.1382 | No |
| 46 | Coq8a | 8645 | 0.043 | 0.1353 | No |
| 47 | Kmt2d | 8657 | 0.042 | 0.1367 | No |
| 48 | Ptprj | 8678 | 0.042 | 0.1376 | No |
| 49 | Asb7 | 8790 | 0.038 | 0.1337 | No |
| 50 | Nrip2 | 8951 | 0.032 | 0.1271 | No |
| 51 | Mx2 | 8994 | 0.031 | 0.1263 | No |
| 52 | Prodh | 9074 | 0.028 | 0.1236 | No |
| 53 | Ryr1 | 9169 | 0.025 | 0.1199 | No |
| 54 | Sphk2 | 9397 | 0.018 | 0.1092 | No |
| 55 | Slc16a7 | 9574 | 0.012 | 0.1008 | No |
| 56 | Zbtb16 | 9615 | 0.011 | 0.0993 | No |
| 57 | Thnsl2 | 9672 | 0.009 | 0.0969 | No |
| 58 | Lfng | 9678 | 0.009 | 0.0970 | No |
| 59 | Mast3 | 9695 | 0.008 | 0.0966 | No |
| 60 | Serpinb2 | 10144 | -0.002 | 0.0739 | No |
| 61 | Thrb | 10182 | -0.003 | 0.0722 | No |
| 62 | Arpp21 | 10587 | -0.016 | 0.0524 | No |
| 63 | Kcnn1 | 10687 | -0.019 | 0.0482 | No |
| 64 | Bard1 | 10717 | -0.020 | 0.0476 | No |
| 65 | Egf | 10899 | -0.025 | 0.0396 | No |
| 66 | Tex15 | 11411 | -0.041 | 0.0155 | No |
| 67 | Pdk2 | 11655 | -0.049 | 0.0054 | No |
| 68 | Plag1 | 12182 | -0.066 | -0.0183 | No |
| 69 | Slc25a23 | 12636 | -0.078 | -0.0378 | No |
| 70 | Stag3 | 12660 | -0.079 | -0.0353 | No |
| 71 | Selenop | 13101 | -0.094 | -0.0534 | No |
| 72 | Btg2 | 13115 | -0.095 | -0.0497 | No |
| 73 | Gamt | 13288 | -0.102 | -0.0538 | No |
| 74 | Ccdc106 | 13317 | -0.102 | -0.0505 | No |
| 75 | Dcc | 13378 | -0.105 | -0.0488 | No |
| 76 | Tgfb2 | 13460 | -0.108 | -0.0480 | No |
| 77 | Myo15a | 13996 | -0.128 | -0.0693 | No |
| 78 | Snn | 14523 | -0.148 | -0.0893 | No |
| 79 | Synpo | 14720 | -0.157 | -0.0921 | No |
| 80 | Vps50 | 14870 | -0.165 | -0.0921 | No |
| 81 | Brdt | 14949 | -0.168 | -0.0884 | No |
| 82 | Cyp39a1 | 15413 | -0.185 | -0.1035 | No |
| 83 | Arhgdig | 15605 | -0.193 | -0.1044 | No |
| 84 | Edar | 15623 | -0.193 | -0.0964 | No |
| 85 | Sgk1 | 15647 | -0.195 | -0.0887 | No |
| 86 | Copz2 | 15943 | -0.211 | -0.0940 | No |
| 87 | Ryr2 | 16023 | -0.214 | -0.0882 | No |
| 88 | Cpb1 | 16139 | -0.221 | -0.0839 | No |
| 89 | Mfsd6 | 16227 | -0.225 | -0.0781 | No |
| 90 | Tnni3 | 16393 | -0.233 | -0.0758 | No |
| 91 | Bmpr1b | 16436 | -0.236 | -0.0671 | No |
| 92 | Hc | 16535 | -0.242 | -0.0610 | No |
| 93 | Hsd11b2 | 16920 | -0.267 | -0.0683 | No |
| 94 | Tgm1 | 17422 | -0.300 | -0.0800 | No |
| 95 | Efhd1 | 18151 | -0.362 | -0.1005 | No |
| 96 | Ybx2 | 18198 | -0.368 | -0.0860 | No |
| 97 | Cpeb3 | 18245 | -0.373 | -0.0713 | No |
| 98 | Rsad2 | 18259 | -0.375 | -0.0548 | No |
| 99 | Kcnd1 | 18330 | -0.383 | -0.0408 | No |
| 100 | Klk8 | 18969 | -0.468 | -0.0518 | No |
| 101 | Itgb1bp2 | 19317 | -0.547 | -0.0445 | No |
| 102 | Mthfr | 19484 | -0.605 | -0.0252 | No |
| 103 | Gp1ba | 19728 | -0.869 | 0.0022 | No |