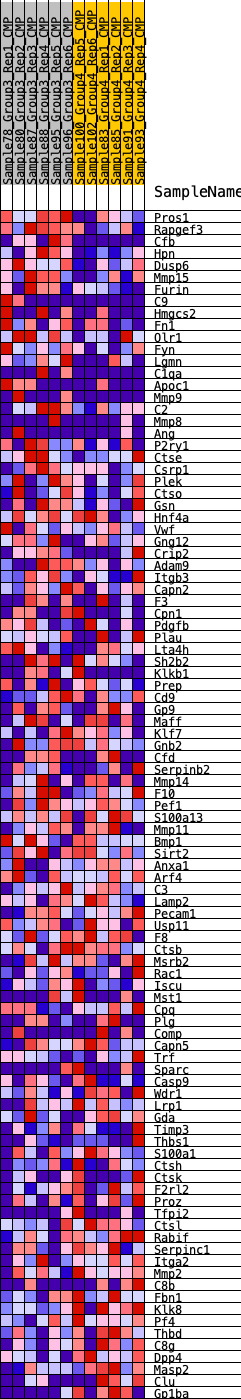
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group3_versus_Group4.CMP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
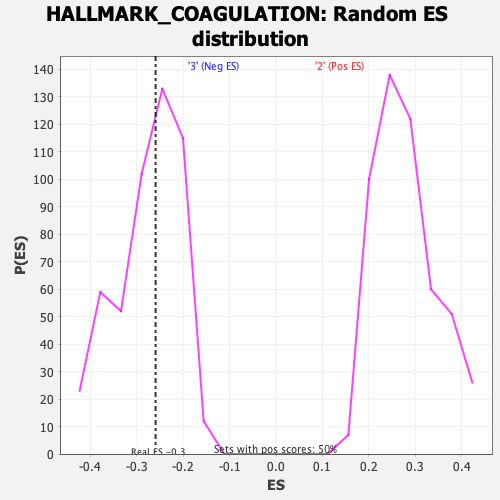
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.25919738 |
| Normalized Enrichment Score (NES) | -0.94078076 |
| Nominal p-value | 0.52620965 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pros1 | 121 | 0.822 | 0.0266 | No |
| 2 | Rapgef3 | 153 | 0.799 | 0.0568 | No |
| 3 | Cfb | 197 | 0.753 | 0.0846 | No |
| 4 | Hpn | 540 | 0.587 | 0.0906 | No |
| 5 | Dusp6 | 580 | 0.577 | 0.1115 | No |
| 6 | Mmp15 | 877 | 0.509 | 0.1167 | No |
| 7 | Furin | 1031 | 0.480 | 0.1281 | No |
| 8 | C9 | 1304 | 0.443 | 0.1319 | No |
| 9 | Hmgcs2 | 1359 | 0.437 | 0.1466 | No |
| 10 | Fn1 | 1694 | 0.398 | 0.1454 | No |
| 11 | Olr1 | 1768 | 0.390 | 0.1572 | No |
| 12 | Fyn | 1989 | 0.369 | 0.1607 | No |
| 13 | Lgmn | 2059 | 0.363 | 0.1717 | No |
| 14 | C1qa | 2312 | 0.340 | 0.1724 | No |
| 15 | Apoc1 | 2374 | 0.336 | 0.1827 | No |
| 16 | Mmp9 | 2486 | 0.327 | 0.1900 | No |
| 17 | C2 | 2535 | 0.325 | 0.2005 | No |
| 18 | Mmp8 | 2744 | 0.307 | 0.2022 | No |
| 19 | Ang | 3020 | 0.286 | 0.1996 | No |
| 20 | P2ry1 | 3247 | 0.268 | 0.1988 | No |
| 21 | Ctse | 3264 | 0.267 | 0.2086 | No |
| 22 | Csrp1 | 4281 | 0.211 | 0.1653 | No |
| 23 | Plek | 4470 | 0.203 | 0.1638 | No |
| 24 | Ctso | 4513 | 0.200 | 0.1696 | No |
| 25 | Gsn | 4649 | 0.194 | 0.1705 | No |
| 26 | Hnf4a | 4679 | 0.192 | 0.1767 | No |
| 27 | Vwf | 4984 | 0.176 | 0.1683 | No |
| 28 | Gng12 | 5059 | 0.173 | 0.1714 | No |
| 29 | Crip2 | 5229 | 0.166 | 0.1694 | No |
| 30 | Adam9 | 5627 | 0.149 | 0.1551 | No |
| 31 | Itgb3 | 6150 | 0.132 | 0.1338 | No |
| 32 | Capn2 | 6412 | 0.122 | 0.1254 | No |
| 33 | F3 | 6891 | 0.103 | 0.1052 | No |
| 34 | Cpn1 | 7011 | 0.099 | 0.1031 | No |
| 35 | Pdgfb | 7325 | 0.088 | 0.0907 | No |
| 36 | Plau | 7841 | 0.070 | 0.0673 | No |
| 37 | Lta4h | 8813 | 0.037 | 0.0195 | No |
| 38 | Sh2b2 | 8899 | 0.035 | 0.0165 | No |
| 39 | Klkb1 | 8906 | 0.034 | 0.0176 | No |
| 40 | Prep | 8943 | 0.033 | 0.0170 | No |
| 41 | Cd9 | 9196 | 0.024 | 0.0052 | No |
| 42 | Gp9 | 9215 | 0.023 | 0.0052 | No |
| 43 | Maff | 9241 | 0.023 | 0.0048 | No |
| 44 | Klf7 | 9314 | 0.020 | 0.0020 | No |
| 45 | Gnb2 | 9635 | 0.010 | -0.0139 | No |
| 46 | Cfd | 9924 | 0.001 | -0.0285 | No |
| 47 | Serpinb2 | 10144 | -0.002 | -0.0395 | No |
| 48 | Mmp14 | 10277 | -0.006 | -0.0460 | No |
| 49 | F10 | 10590 | -0.016 | -0.0612 | No |
| 50 | Pef1 | 10811 | -0.023 | -0.0715 | No |
| 51 | S100a13 | 10828 | -0.023 | -0.0714 | No |
| 52 | Mmp11 | 11185 | -0.035 | -0.0881 | No |
| 53 | Bmp1 | 11344 | -0.039 | -0.0946 | No |
| 54 | Sirt2 | 11391 | -0.041 | -0.0953 | No |
| 55 | Anxa1 | 12187 | -0.066 | -0.1331 | No |
| 56 | Arf4 | 12276 | -0.069 | -0.1348 | No |
| 57 | C3 | 12398 | -0.071 | -0.1381 | No |
| 58 | Lamp2 | 12519 | -0.075 | -0.1412 | No |
| 59 | Pecam1 | 12588 | -0.077 | -0.1416 | No |
| 60 | Usp11 | 12856 | -0.085 | -0.1518 | No |
| 61 | F8 | 13076 | -0.094 | -0.1592 | No |
| 62 | Ctsb | 13077 | -0.094 | -0.1555 | No |
| 63 | Msrb2 | 13081 | -0.094 | -0.1519 | No |
| 64 | Rac1 | 13144 | -0.096 | -0.1512 | No |
| 65 | Iscu | 13522 | -0.110 | -0.1660 | No |
| 66 | Mst1 | 13636 | -0.114 | -0.1672 | No |
| 67 | Cpq | 14268 | -0.139 | -0.1937 | No |
| 68 | Plg | 14360 | -0.142 | -0.1927 | No |
| 69 | Comp | 14625 | -0.153 | -0.2000 | No |
| 70 | Capn5 | 15639 | -0.194 | -0.2438 | No |
| 71 | Trf | 15726 | -0.199 | -0.2402 | No |
| 72 | Sparc | 15788 | -0.202 | -0.2353 | No |
| 73 | Casp9 | 16192 | -0.224 | -0.2468 | No |
| 74 | Wdr1 | 16205 | -0.224 | -0.2385 | No |
| 75 | Lrp1 | 16612 | -0.247 | -0.2493 | Yes |
| 76 | Gda | 16788 | -0.259 | -0.2479 | Yes |
| 77 | Timp3 | 16814 | -0.260 | -0.2388 | Yes |
| 78 | Thbs1 | 16889 | -0.265 | -0.2321 | Yes |
| 79 | S100a1 | 17133 | -0.281 | -0.2332 | Yes |
| 80 | Ctsh | 17271 | -0.291 | -0.2286 | Yes |
| 81 | Ctsk | 17544 | -0.309 | -0.2301 | Yes |
| 82 | F2rl2 | 18117 | -0.358 | -0.2449 | Yes |
| 83 | Proz | 18258 | -0.375 | -0.2372 | Yes |
| 84 | Tfpi2 | 18318 | -0.382 | -0.2250 | Yes |
| 85 | Ctsl | 18343 | -0.384 | -0.2109 | Yes |
| 86 | Rabif | 18609 | -0.412 | -0.2079 | Yes |
| 87 | Serpinc1 | 18629 | -0.415 | -0.1924 | Yes |
| 88 | Itga2 | 18645 | -0.417 | -0.1766 | Yes |
| 89 | Mmp2 | 18797 | -0.438 | -0.1668 | Yes |
| 90 | C8b | 18884 | -0.451 | -0.1532 | Yes |
| 91 | Fbn1 | 18919 | -0.457 | -0.1367 | Yes |
| 92 | Klk8 | 18969 | -0.468 | -0.1206 | Yes |
| 93 | Pf4 | 18983 | -0.471 | -0.1025 | Yes |
| 94 | Thbd | 18987 | -0.472 | -0.0839 | Yes |
| 95 | C8g | 19107 | -0.493 | -0.0703 | Yes |
| 96 | Dpp4 | 19138 | -0.500 | -0.0520 | Yes |
| 97 | Masp2 | 19504 | -0.616 | -0.0460 | Yes |
| 98 | Clu | 19523 | -0.626 | -0.0220 | Yes |
| 99 | Gp1ba | 19728 | -0.869 | 0.0022 | Yes |