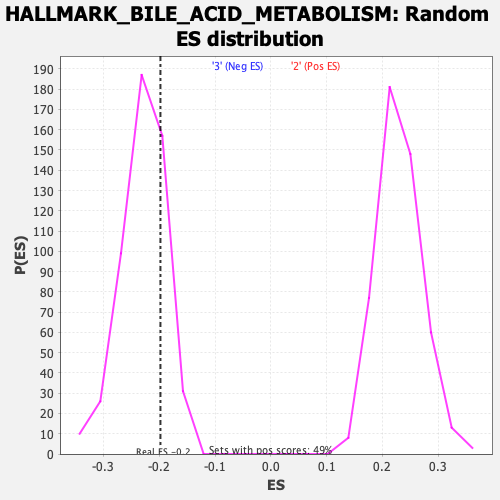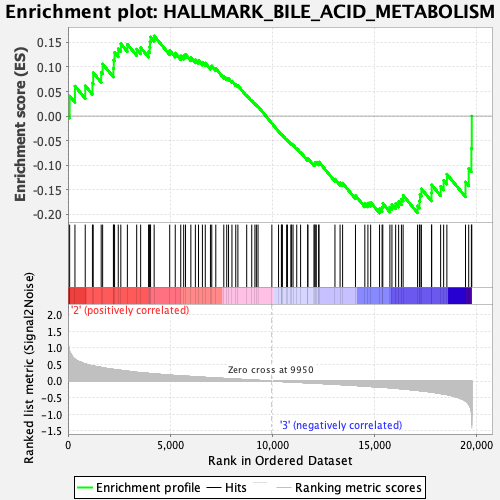
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group3_versus_Group4.CMP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.19776055 |
| Normalized Enrichment Score (NES) | -0.8639894 |
| Nominal p-value | 0.77843136 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc27a2 | 81 | 0.880 | 0.0408 | No |
| 2 | Slc22a18 | 340 | 0.655 | 0.0611 | No |
| 3 | Dhcr24 | 844 | 0.514 | 0.0618 | No |
| 4 | Ar | 1196 | 0.456 | 0.0672 | No |
| 5 | Nr0b2 | 1234 | 0.452 | 0.0884 | No |
| 6 | Nudt12 | 1625 | 0.409 | 0.0894 | No |
| 7 | Gnpat | 1696 | 0.398 | 0.1062 | No |
| 8 | Gnmt | 2223 | 0.349 | 0.0972 | No |
| 9 | Mlycd | 2241 | 0.347 | 0.1141 | No |
| 10 | Fdxr | 2280 | 0.342 | 0.1296 | No |
| 11 | Tfcp2l1 | 2461 | 0.329 | 0.1373 | No |
| 12 | Acsl1 | 2586 | 0.321 | 0.1474 | No |
| 13 | Pex11a | 2907 | 0.296 | 0.1462 | No |
| 14 | Klf1 | 3363 | 0.261 | 0.1364 | No |
| 15 | Pex19 | 3557 | 0.250 | 0.1393 | No |
| 16 | Pnpla8 | 3944 | 0.230 | 0.1315 | No |
| 17 | Akr1d1 | 3995 | 0.227 | 0.1405 | No |
| 18 | Pecr | 4014 | 0.226 | 0.1511 | No |
| 19 | Cyp7b1 | 4042 | 0.225 | 0.1612 | No |
| 20 | Nedd4 | 4218 | 0.215 | 0.1633 | No |
| 21 | Ephx2 | 4980 | 0.177 | 0.1336 | No |
| 22 | Abca2 | 5251 | 0.166 | 0.1284 | No |
| 23 | Bmp6 | 5520 | 0.154 | 0.1226 | No |
| 24 | Acsl5 | 5664 | 0.147 | 0.1229 | No |
| 25 | Gclm | 5754 | 0.144 | 0.1257 | No |
| 26 | Abca4 | 6015 | 0.137 | 0.1195 | No |
| 27 | Pex1 | 6236 | 0.128 | 0.1148 | No |
| 28 | Prdx5 | 6390 | 0.123 | 0.1133 | No |
| 29 | Pxmp2 | 6581 | 0.115 | 0.1095 | No |
| 30 | Pex6 | 6721 | 0.109 | 0.1080 | No |
| 31 | Idh2 | 6974 | 0.101 | 0.1004 | No |
| 32 | Lonp2 | 7039 | 0.098 | 0.1021 | No |
| 33 | Pex16 | 7236 | 0.091 | 0.0968 | No |
| 34 | Slc23a2 | 7628 | 0.077 | 0.0809 | No |
| 35 | Abca6 | 7771 | 0.071 | 0.0773 | No |
| 36 | Pex26 | 7860 | 0.070 | 0.0764 | No |
| 37 | Npc1 | 8027 | 0.064 | 0.0712 | No |
| 38 | Abca9 | 8213 | 0.058 | 0.0647 | No |
| 39 | Abcg4 | 8318 | 0.054 | 0.0622 | No |
| 40 | Cat | 8749 | 0.040 | 0.0424 | No |
| 41 | Dio2 | 8996 | 0.031 | 0.0315 | No |
| 42 | Slc23a1 | 9152 | 0.025 | 0.0249 | No |
| 43 | Sod1 | 9223 | 0.023 | 0.0225 | No |
| 44 | Nr3c2 | 9304 | 0.021 | 0.0195 | No |
| 45 | Gc | 9981 | 0.000 | -0.0149 | No |
| 46 | Lipe | 10308 | -0.007 | -0.0311 | No |
| 47 | Aldh9a1 | 10448 | -0.012 | -0.0375 | No |
| 48 | Idh1 | 10471 | -0.012 | -0.0380 | No |
| 49 | Phyh | 10495 | -0.013 | -0.0385 | No |
| 50 | Hsd3b7 | 10704 | -0.019 | -0.0481 | No |
| 51 | Optn | 10745 | -0.021 | -0.0491 | No |
| 52 | Pex13 | 10911 | -0.025 | -0.0562 | No |
| 53 | Pex7 | 10949 | -0.027 | -0.0567 | No |
| 54 | Atxn1 | 11019 | -0.029 | -0.0587 | No |
| 55 | Abcd1 | 11199 | -0.035 | -0.0660 | No |
| 56 | Aldh1a1 | 11388 | -0.041 | -0.0735 | No |
| 57 | Scp2 | 11737 | -0.052 | -0.0885 | No |
| 58 | Isoc1 | 11743 | -0.052 | -0.0861 | No |
| 59 | Nr1h4 | 12043 | -0.062 | -0.0981 | No |
| 60 | Hacl1 | 12092 | -0.063 | -0.0974 | No |
| 61 | Lck | 12097 | -0.063 | -0.0943 | No |
| 62 | Pfkm | 12156 | -0.066 | -0.0939 | No |
| 63 | Abcd2 | 12270 | -0.069 | -0.0961 | No |
| 64 | Pipox | 12291 | -0.070 | -0.0936 | No |
| 65 | Cyp46a1 | 13070 | -0.094 | -0.1283 | No |
| 66 | Retsat | 13321 | -0.103 | -0.1358 | No |
| 67 | Abca3 | 13445 | -0.107 | -0.1366 | No |
| 68 | Hsd17b11 | 14075 | -0.131 | -0.1619 | No |
| 69 | Pex11g | 14533 | -0.149 | -0.1775 | No |
| 70 | Rbp1 | 14684 | -0.155 | -0.1772 | No |
| 71 | Idi1 | 14818 | -0.162 | -0.1757 | No |
| 72 | Aqp9 | 15253 | -0.177 | -0.1887 | Yes |
| 73 | Fads1 | 15378 | -0.183 | -0.1857 | Yes |
| 74 | Cyp39a1 | 15413 | -0.185 | -0.1780 | Yes |
| 75 | Sult2b1 | 15760 | -0.201 | -0.1853 | Yes |
| 76 | Slc35b2 | 15860 | -0.206 | -0.1798 | Yes |
| 77 | Fads2 | 16042 | -0.215 | -0.1781 | Yes |
| 78 | Amacr | 16188 | -0.224 | -0.1740 | Yes |
| 79 | Hsd17b4 | 16325 | -0.231 | -0.1692 | Yes |
| 80 | Bcar3 | 16408 | -0.234 | -0.1614 | Yes |
| 81 | Crot | 17115 | -0.280 | -0.1830 | Yes |
| 82 | Abca5 | 17206 | -0.286 | -0.1729 | Yes |
| 83 | Abcd3 | 17235 | -0.289 | -0.1596 | Yes |
| 84 | Paox | 17304 | -0.294 | -0.1481 | Yes |
| 85 | Pex12 | 17796 | -0.327 | -0.1563 | Yes |
| 86 | Gstk1 | 17808 | -0.329 | -0.1401 | Yes |
| 87 | Slc29a1 | 18244 | -0.373 | -0.1432 | Yes |
| 88 | Efhc1 | 18392 | -0.388 | -0.1309 | Yes |
| 89 | Cyp27a1 | 18550 | -0.403 | -0.1183 | Yes |
| 90 | Abca8b | 19462 | -0.597 | -0.1341 | Yes |
| 91 | Rxra | 19619 | -0.688 | -0.1070 | Yes |
| 92 | Soat2 | 19750 | -0.936 | -0.0658 | Yes |
| 93 | Abca1 | 19769 | -1.310 | 0.0001 | Yes |