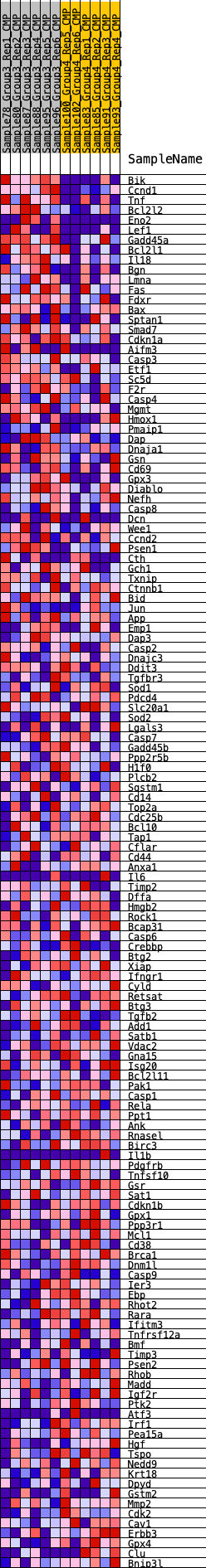
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group3_versus_Group4.CMP_Pheno.cls#Group3_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
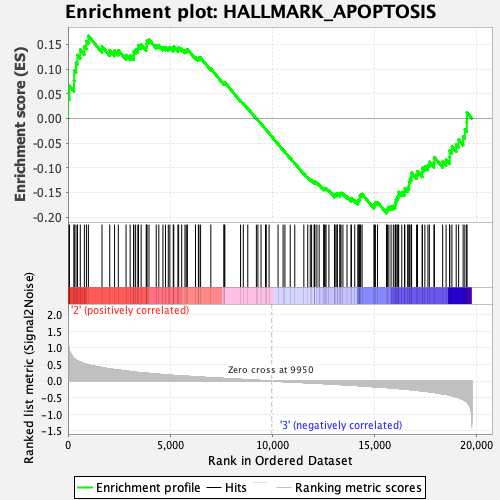
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.19201699 |
| Normalized Enrichment Score (NES) | -0.9031616 |
| Nominal p-value | 0.66536963 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bik | 6 | 1.315 | 0.0413 | No |
| 2 | Ccnd1 | 71 | 0.893 | 0.0662 | No |
| 3 | Tnf | 293 | 0.678 | 0.0764 | No |
| 4 | Bcl2l2 | 304 | 0.673 | 0.0972 | No |
| 5 | Eno2 | 393 | 0.634 | 0.1127 | No |
| 6 | Lef1 | 464 | 0.609 | 0.1284 | No |
| 7 | Gadd45a | 601 | 0.571 | 0.1395 | No |
| 8 | Bcl2l1 | 801 | 0.524 | 0.1459 | No |
| 9 | Il18 | 903 | 0.505 | 0.1567 | No |
| 10 | Bgn | 998 | 0.487 | 0.1673 | No |
| 11 | Lmna | 1664 | 0.402 | 0.1462 | No |
| 12 | Fas | 2043 | 0.365 | 0.1384 | No |
| 13 | Fdxr | 2280 | 0.342 | 0.1372 | No |
| 14 | Bax | 2463 | 0.328 | 0.1383 | No |
| 15 | Sptan1 | 2843 | 0.301 | 0.1285 | No |
| 16 | Smad7 | 3043 | 0.284 | 0.1274 | No |
| 17 | Cdkn1a | 3213 | 0.271 | 0.1273 | No |
| 18 | Aifm3 | 3220 | 0.270 | 0.1356 | No |
| 19 | Casp3 | 3311 | 0.264 | 0.1393 | No |
| 20 | Etf1 | 3422 | 0.258 | 0.1419 | No |
| 21 | Sc5d | 3450 | 0.256 | 0.1486 | No |
| 22 | F2r | 3581 | 0.248 | 0.1498 | No |
| 23 | Casp4 | 3829 | 0.237 | 0.1447 | No |
| 24 | Mgmt | 3856 | 0.235 | 0.1508 | No |
| 25 | Hmox1 | 3864 | 0.234 | 0.1579 | No |
| 26 | Pmaip1 | 3964 | 0.229 | 0.1601 | No |
| 27 | Dap | 4319 | 0.209 | 0.1486 | No |
| 28 | Dnaja1 | 4446 | 0.204 | 0.1487 | No |
| 29 | Gsn | 4649 | 0.194 | 0.1445 | No |
| 30 | Cd69 | 4773 | 0.187 | 0.1442 | No |
| 31 | Gpx3 | 4910 | 0.180 | 0.1429 | No |
| 32 | Diablo | 4988 | 0.176 | 0.1446 | No |
| 33 | Nefh | 5153 | 0.169 | 0.1415 | No |
| 34 | Casp8 | 5180 | 0.168 | 0.1455 | No |
| 35 | Dcn | 5382 | 0.160 | 0.1403 | No |
| 36 | Wee1 | 5418 | 0.158 | 0.1436 | No |
| 37 | Ccnd2 | 5566 | 0.152 | 0.1409 | No |
| 38 | Psen1 | 5727 | 0.145 | 0.1373 | No |
| 39 | Cth | 5791 | 0.143 | 0.1386 | No |
| 40 | Gch1 | 5845 | 0.140 | 0.1403 | No |
| 41 | Txnip | 6243 | 0.128 | 0.1241 | No |
| 42 | Ctnnb1 | 6384 | 0.123 | 0.1209 | No |
| 43 | Bid | 6410 | 0.122 | 0.1235 | No |
| 44 | Jun | 6492 | 0.118 | 0.1231 | No |
| 45 | App | 6993 | 0.100 | 0.1008 | No |
| 46 | Emp1 | 7631 | 0.077 | 0.0708 | No |
| 47 | Dap3 | 7652 | 0.076 | 0.0722 | No |
| 48 | Casp2 | 7679 | 0.076 | 0.0732 | No |
| 49 | Dnajc3 | 8449 | 0.050 | 0.0356 | No |
| 50 | Ddit3 | 8583 | 0.045 | 0.0303 | No |
| 51 | Tgfbr3 | 8802 | 0.038 | 0.0204 | No |
| 52 | Sod1 | 9223 | 0.023 | -0.0003 | No |
| 53 | Pdcd4 | 9298 | 0.021 | -0.0034 | No |
| 54 | Slc20a1 | 9450 | 0.017 | -0.0106 | No |
| 55 | Sod2 | 9670 | 0.009 | -0.0215 | No |
| 56 | Lgals3 | 9708 | 0.008 | -0.0231 | No |
| 57 | Casp7 | 9844 | 0.004 | -0.0299 | No |
| 58 | Gadd45b | 10285 | -0.006 | -0.0521 | No |
| 59 | Ppp2r5b | 10530 | -0.014 | -0.0640 | No |
| 60 | H1f0 | 10620 | -0.017 | -0.0681 | No |
| 61 | Plcb2 | 10880 | -0.025 | -0.0805 | No |
| 62 | Sqstm1 | 11102 | -0.032 | -0.0907 | No |
| 63 | Cd14 | 11547 | -0.045 | -0.1119 | No |
| 64 | Top2a | 11738 | -0.052 | -0.1199 | No |
| 65 | Cdc25b | 11861 | -0.056 | -0.1244 | No |
| 66 | Bcl10 | 11927 | -0.058 | -0.1258 | No |
| 67 | Tap1 | 12050 | -0.062 | -0.1301 | No |
| 68 | Cflar | 12072 | -0.063 | -0.1292 | No |
| 69 | Cd44 | 12090 | -0.063 | -0.1281 | No |
| 70 | Anxa1 | 12187 | -0.066 | -0.1309 | No |
| 71 | Il6 | 12296 | -0.070 | -0.1341 | No |
| 72 | Timp2 | 12517 | -0.075 | -0.1430 | No |
| 73 | Dffa | 12550 | -0.076 | -0.1422 | No |
| 74 | Hmgb2 | 12587 | -0.077 | -0.1416 | No |
| 75 | Rock1 | 12634 | -0.078 | -0.1415 | No |
| 76 | Bcap31 | 12774 | -0.083 | -0.1460 | No |
| 77 | Casp6 | 13046 | -0.093 | -0.1568 | No |
| 78 | Crebbp | 13049 | -0.093 | -0.1540 | No |
| 79 | Btg2 | 13115 | -0.095 | -0.1543 | No |
| 80 | Xiap | 13177 | -0.097 | -0.1544 | No |
| 81 | Ifngr1 | 13182 | -0.098 | -0.1515 | No |
| 82 | Cyld | 13312 | -0.102 | -0.1548 | No |
| 83 | Retsat | 13321 | -0.103 | -0.1520 | No |
| 84 | Btg3 | 13367 | -0.104 | -0.1510 | No |
| 85 | Tgfb2 | 13460 | -0.108 | -0.1523 | No |
| 86 | Add1 | 13661 | -0.115 | -0.1588 | No |
| 87 | Satb1 | 13856 | -0.123 | -0.1648 | No |
| 88 | Vdac2 | 13875 | -0.123 | -0.1619 | No |
| 89 | Gna15 | 14040 | -0.129 | -0.1661 | No |
| 90 | Isg20 | 14197 | -0.136 | -0.1698 | No |
| 91 | Bcl2l11 | 14206 | -0.137 | -0.1659 | No |
| 92 | Pak1 | 14253 | -0.139 | -0.1638 | No |
| 93 | Casp1 | 14292 | -0.140 | -0.1613 | No |
| 94 | Rela | 14293 | -0.140 | -0.1569 | No |
| 95 | Ppt1 | 14331 | -0.141 | -0.1543 | No |
| 96 | Ank | 14399 | -0.143 | -0.1532 | No |
| 97 | Rnasel | 14979 | -0.170 | -0.1773 | No |
| 98 | Birc3 | 15011 | -0.171 | -0.1735 | No |
| 99 | Il1b | 15050 | -0.172 | -0.1700 | No |
| 100 | Pdgfrb | 15155 | -0.173 | -0.1698 | No |
| 101 | Tnfsf10 | 15592 | -0.192 | -0.1859 | Yes |
| 102 | Gsr | 15646 | -0.195 | -0.1825 | Yes |
| 103 | Sat1 | 15711 | -0.198 | -0.1795 | Yes |
| 104 | Cdkn1b | 15818 | -0.204 | -0.1785 | Yes |
| 105 | Gpx1 | 15928 | -0.209 | -0.1774 | Yes |
| 106 | Ppp3r1 | 16018 | -0.214 | -0.1752 | Yes |
| 107 | Mcl1 | 16028 | -0.214 | -0.1689 | Yes |
| 108 | Cd38 | 16062 | -0.216 | -0.1637 | Yes |
| 109 | Brca1 | 16123 | -0.220 | -0.1598 | Yes |
| 110 | Dnm1l | 16170 | -0.223 | -0.1551 | Yes |
| 111 | Casp9 | 16192 | -0.224 | -0.1491 | Yes |
| 112 | Ier3 | 16342 | -0.232 | -0.1494 | Yes |
| 113 | Ebp | 16477 | -0.239 | -0.1486 | Yes |
| 114 | Rhot2 | 16490 | -0.240 | -0.1417 | Yes |
| 115 | Rara | 16630 | -0.248 | -0.1409 | Yes |
| 116 | Ifitm3 | 16696 | -0.253 | -0.1362 | Yes |
| 117 | Tnfrsf12a | 16699 | -0.253 | -0.1283 | Yes |
| 118 | Bmf | 16745 | -0.255 | -0.1226 | Yes |
| 119 | Timp3 | 16814 | -0.260 | -0.1178 | Yes |
| 120 | Psen2 | 16822 | -0.261 | -0.1099 | Yes |
| 121 | Rhob | 17067 | -0.276 | -0.1136 | Yes |
| 122 | Madd | 17112 | -0.280 | -0.1070 | Yes |
| 123 | Igf2r | 17343 | -0.296 | -0.1094 | Yes |
| 124 | Ptk2 | 17348 | -0.297 | -0.1002 | Yes |
| 125 | Atf3 | 17473 | -0.305 | -0.0969 | Yes |
| 126 | Irf1 | 17623 | -0.315 | -0.0946 | Yes |
| 127 | Pea15a | 17691 | -0.319 | -0.0879 | Yes |
| 128 | Hgf | 17919 | -0.339 | -0.0887 | Yes |
| 129 | Tspo | 17938 | -0.341 | -0.0789 | Yes |
| 130 | Nedd9 | 18344 | -0.384 | -0.0874 | Yes |
| 131 | Krt18 | 18512 | -0.399 | -0.0833 | Yes |
| 132 | Dpyd | 18685 | -0.422 | -0.0787 | Yes |
| 133 | Gstm2 | 18691 | -0.423 | -0.0656 | Yes |
| 134 | Mmp2 | 18797 | -0.438 | -0.0571 | Yes |
| 135 | Cdk2 | 19007 | -0.475 | -0.0527 | Yes |
| 136 | Cav1 | 19125 | -0.497 | -0.0430 | Yes |
| 137 | Erbb3 | 19346 | -0.555 | -0.0367 | Yes |
| 138 | Gpx4 | 19430 | -0.583 | -0.0225 | Yes |
| 139 | Clu | 19523 | -0.626 | -0.0074 | Yes |
| 140 | Bnip3l | 19531 | -0.631 | 0.0122 | Yes |