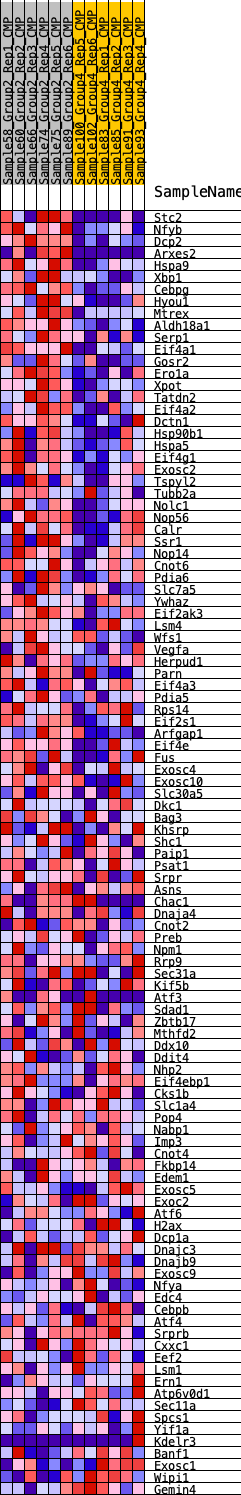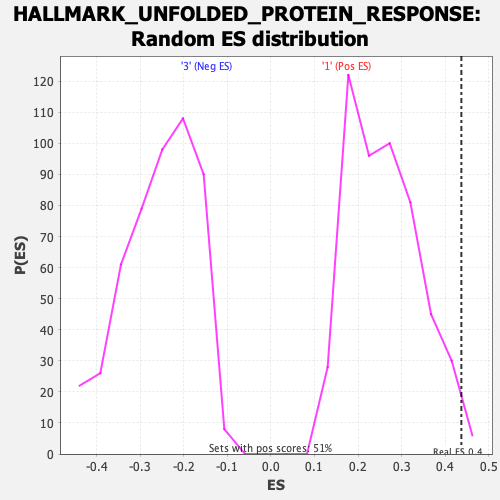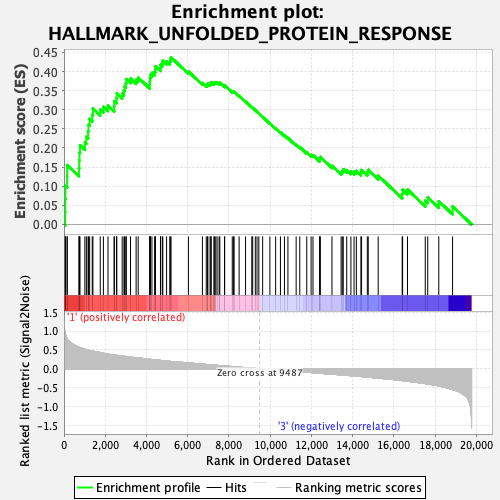
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group4.CMP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.43705434 |
| Normalized Enrichment Score (NES) | 1.6862283 |
| Nominal p-value | 0.011811024 |
| FDR q-value | 0.06690403 |
| FWER p-Value | 0.155 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Stc2 | 60 | 0.944 | 0.0320 | Yes |
| 2 | Nfyb | 63 | 0.936 | 0.0666 | Yes |
| 3 | Dcp2 | 70 | 0.920 | 0.1005 | Yes |
| 4 | Arxes2 | 151 | 0.794 | 0.1259 | Yes |
| 5 | Hspa9 | 154 | 0.791 | 0.1551 | Yes |
| 6 | Xbp1 | 725 | 0.567 | 0.1472 | Yes |
| 7 | Cebpg | 740 | 0.565 | 0.1675 | Yes |
| 8 | Hyou1 | 751 | 0.562 | 0.1878 | Yes |
| 9 | Mtrex | 775 | 0.558 | 0.2074 | Yes |
| 10 | Aldh18a1 | 1012 | 0.515 | 0.2145 | Yes |
| 11 | Serp1 | 1085 | 0.503 | 0.2295 | Yes |
| 12 | Eif4a1 | 1168 | 0.493 | 0.2436 | Yes |
| 13 | Gosr2 | 1186 | 0.491 | 0.2610 | Yes |
| 14 | Ero1a | 1243 | 0.484 | 0.2761 | Yes |
| 15 | Xpot | 1369 | 0.469 | 0.2872 | Yes |
| 16 | Tatdn2 | 1403 | 0.465 | 0.3027 | Yes |
| 17 | Eif4a2 | 1759 | 0.427 | 0.3005 | Yes |
| 18 | Dctn1 | 1911 | 0.409 | 0.3080 | Yes |
| 19 | Hsp90b1 | 2134 | 0.387 | 0.3111 | Yes |
| 20 | Hspa5 | 2434 | 0.362 | 0.3093 | Yes |
| 21 | Eif4g1 | 2438 | 0.361 | 0.3226 | Yes |
| 22 | Exosc2 | 2551 | 0.355 | 0.3301 | Yes |
| 23 | Tspyl2 | 2558 | 0.354 | 0.3429 | Yes |
| 24 | Tubb2a | 2822 | 0.336 | 0.3420 | Yes |
| 25 | Nolc1 | 2908 | 0.329 | 0.3499 | Yes |
| 26 | Nop56 | 2937 | 0.328 | 0.3607 | Yes |
| 27 | Calr | 2998 | 0.324 | 0.3696 | Yes |
| 28 | Ssr1 | 3023 | 0.322 | 0.3804 | Yes |
| 29 | Nop14 | 3226 | 0.309 | 0.3816 | Yes |
| 30 | Cnot6 | 3498 | 0.291 | 0.3786 | Yes |
| 31 | Pdia6 | 3598 | 0.285 | 0.3841 | Yes |
| 32 | Slc7a5 | 4156 | 0.250 | 0.3651 | Yes |
| 33 | Ywhaz | 4165 | 0.249 | 0.3739 | Yes |
| 34 | Eif2ak3 | 4171 | 0.249 | 0.3829 | Yes |
| 35 | Lsm4 | 4193 | 0.248 | 0.3911 | Yes |
| 36 | Wfs1 | 4265 | 0.242 | 0.3964 | Yes |
| 37 | Vegfa | 4396 | 0.235 | 0.3986 | Yes |
| 38 | Herpud1 | 4428 | 0.233 | 0.4056 | Yes |
| 39 | Parn | 4439 | 0.232 | 0.4138 | Yes |
| 40 | Eif4a3 | 4679 | 0.220 | 0.4098 | Yes |
| 41 | Pdia5 | 4683 | 0.220 | 0.4178 | Yes |
| 42 | Rps14 | 4767 | 0.214 | 0.4215 | Yes |
| 43 | Eif2s1 | 4792 | 0.212 | 0.4282 | Yes |
| 44 | Arfgap1 | 4976 | 0.202 | 0.4264 | Yes |
| 45 | Eif4e | 5133 | 0.195 | 0.4257 | Yes |
| 46 | Fus | 5156 | 0.193 | 0.4317 | Yes |
| 47 | Exosc4 | 5192 | 0.191 | 0.4371 | Yes |
| 48 | Exosc10 | 6034 | 0.153 | 0.4000 | No |
| 49 | Slc30a5 | 6713 | 0.121 | 0.3700 | No |
| 50 | Dkc1 | 6899 | 0.112 | 0.3647 | No |
| 51 | Bag3 | 6962 | 0.108 | 0.3656 | No |
| 52 | Khsrp | 6978 | 0.108 | 0.3688 | No |
| 53 | Shc1 | 7085 | 0.103 | 0.3673 | No |
| 54 | Paip1 | 7125 | 0.101 | 0.3690 | No |
| 55 | Psat1 | 7135 | 0.101 | 0.3723 | No |
| 56 | Srpr | 7273 | 0.095 | 0.3689 | No |
| 57 | Asns | 7294 | 0.093 | 0.3713 | No |
| 58 | Chac1 | 7360 | 0.090 | 0.3714 | No |
| 59 | Dnaja4 | 7425 | 0.088 | 0.3714 | No |
| 60 | Cnot2 | 7522 | 0.083 | 0.3696 | No |
| 61 | Preb | 7557 | 0.082 | 0.3709 | No |
| 62 | Npm1 | 7787 | 0.072 | 0.3619 | No |
| 63 | Rrp9 | 7792 | 0.072 | 0.3644 | No |
| 64 | Sec31a | 8161 | 0.055 | 0.3477 | No |
| 65 | Kif5b | 8225 | 0.052 | 0.3464 | No |
| 66 | Atf3 | 8249 | 0.051 | 0.3471 | No |
| 67 | Sdad1 | 8494 | 0.040 | 0.3362 | No |
| 68 | Zbtb17 | 8806 | 0.027 | 0.3214 | No |
| 69 | Mthfd2 | 9109 | 0.015 | 0.3066 | No |
| 70 | Ddx10 | 9152 | 0.013 | 0.3050 | No |
| 71 | Ddit4 | 9280 | 0.008 | 0.2988 | No |
| 72 | Nhp2 | 9362 | 0.004 | 0.2948 | No |
| 73 | Eif4ebp1 | 9451 | 0.001 | 0.2904 | No |
| 74 | Cks1b | 9635 | -0.004 | 0.2813 | No |
| 75 | Slc1a4 | 9986 | -0.018 | 0.2641 | No |
| 76 | Pop4 | 10265 | -0.030 | 0.2511 | No |
| 77 | Nabp1 | 10498 | -0.040 | 0.2408 | No |
| 78 | Imp3 | 10692 | -0.047 | 0.2327 | No |
| 79 | Cnot4 | 10861 | -0.055 | 0.2262 | No |
| 80 | Fkbp14 | 11260 | -0.070 | 0.2086 | No |
| 81 | Edem1 | 11436 | -0.077 | 0.2025 | No |
| 82 | Exosc5 | 11775 | -0.091 | 0.1887 | No |
| 83 | Exoc2 | 11985 | -0.100 | 0.1818 | No |
| 84 | Atf6 | 12076 | -0.105 | 0.1812 | No |
| 85 | H2ax | 12401 | -0.119 | 0.1691 | No |
| 86 | Dcp1a | 12403 | -0.119 | 0.1735 | No |
| 87 | Dnajc3 | 12432 | -0.121 | 0.1765 | No |
| 88 | Dnajb9 | 12998 | -0.145 | 0.1532 | No |
| 89 | Exosc9 | 13441 | -0.164 | 0.1368 | No |
| 90 | Nfya | 13498 | -0.167 | 0.1402 | No |
| 91 | Edc4 | 13544 | -0.170 | 0.1442 | No |
| 92 | Cebpb | 13713 | -0.176 | 0.1422 | No |
| 93 | Atf4 | 13916 | -0.184 | 0.1388 | No |
| 94 | Srprb | 14069 | -0.192 | 0.1381 | No |
| 95 | Cxxc1 | 14180 | -0.196 | 0.1398 | No |
| 96 | Eef2 | 14401 | -0.207 | 0.1363 | No |
| 97 | Lsm1 | 14433 | -0.208 | 0.1425 | No |
| 98 | Ern1 | 14712 | -0.222 | 0.1366 | No |
| 99 | Atp6v0d1 | 14756 | -0.225 | 0.1428 | No |
| 100 | Sec11a | 15239 | -0.249 | 0.1275 | No |
| 101 | Spcs1 | 16403 | -0.315 | 0.0800 | No |
| 102 | Yif1a | 16424 | -0.317 | 0.0908 | No |
| 103 | Kdelr3 | 16661 | -0.331 | 0.0911 | No |
| 104 | Banf1 | 17522 | -0.393 | 0.0620 | No |
| 105 | Exosc1 | 17642 | -0.402 | 0.0709 | No |
| 106 | Wipi1 | 18179 | -0.456 | 0.0605 | No |
| 107 | Gemin4 | 18847 | -0.549 | 0.0470 | No |