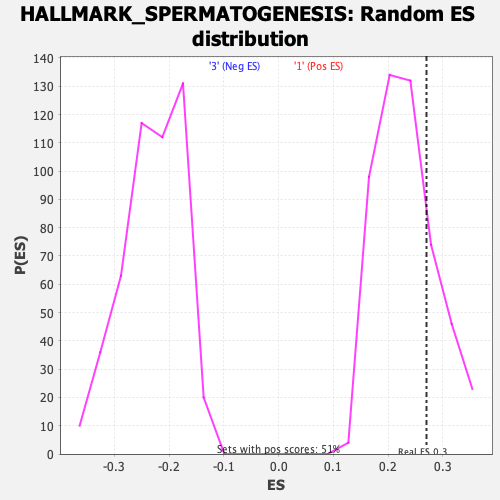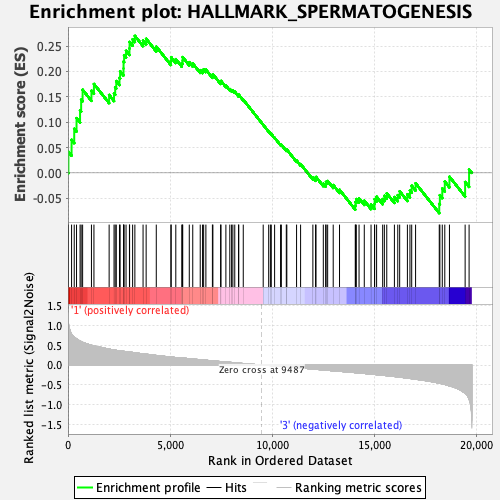
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group4.CMP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.2699949 |
| Normalized Enrichment Score (NES) | 1.1592138 |
| Nominal p-value | 0.22700587 |
| FDR q-value | 0.4620584 |
| FWER p-Value | 0.875 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Jam3 | 20 | 1.057 | 0.0416 | Yes |
| 2 | Aurka | 175 | 0.775 | 0.0651 | Yes |
| 3 | Pgs1 | 305 | 0.703 | 0.0870 | Yes |
| 4 | Nefh | 418 | 0.658 | 0.1078 | Yes |
| 5 | Ncaph | 591 | 0.600 | 0.1233 | Yes |
| 6 | Kif2c | 644 | 0.588 | 0.1444 | Yes |
| 7 | Clgn | 716 | 0.569 | 0.1638 | Yes |
| 8 | Ide | 1148 | 0.496 | 0.1619 | Yes |
| 9 | Gapdhs | 1270 | 0.480 | 0.1752 | Yes |
| 10 | Cdk1 | 2014 | 0.400 | 0.1536 | Yes |
| 11 | Topbp1 | 2254 | 0.376 | 0.1566 | Yes |
| 12 | Hspa4l | 2310 | 0.372 | 0.1688 | Yes |
| 13 | Vdac3 | 2364 | 0.367 | 0.1810 | Yes |
| 14 | Zc2hc1c | 2521 | 0.357 | 0.1874 | Yes |
| 15 | Prkar2a | 2552 | 0.355 | 0.2002 | Yes |
| 16 | Dmc1 | 2718 | 0.343 | 0.2057 | Yes |
| 17 | Scg3 | 2720 | 0.343 | 0.2195 | Yes |
| 18 | Art3 | 2748 | 0.342 | 0.2319 | Yes |
| 19 | Nek2 | 2842 | 0.335 | 0.2407 | Yes |
| 20 | Bub1 | 3009 | 0.323 | 0.2453 | Yes |
| 21 | Hspa2 | 3016 | 0.322 | 0.2580 | Yes |
| 22 | Coil | 3164 | 0.315 | 0.2632 | Yes |
| 23 | Agfg1 | 3274 | 0.304 | 0.2700 | Yes |
| 24 | Tekt2 | 3675 | 0.280 | 0.2610 | No |
| 25 | Ccnb2 | 3826 | 0.272 | 0.2643 | No |
| 26 | Clvs1 | 4322 | 0.239 | 0.2488 | No |
| 27 | Pebp1 | 5038 | 0.199 | 0.2205 | No |
| 28 | Dbf4 | 5051 | 0.198 | 0.2279 | No |
| 29 | Rfc4 | 5278 | 0.189 | 0.2241 | No |
| 30 | Ldhc | 5570 | 0.174 | 0.2163 | No |
| 31 | Nos1 | 5611 | 0.172 | 0.2212 | No |
| 32 | Arl4a | 5613 | 0.172 | 0.2281 | No |
| 33 | Ttk | 5938 | 0.159 | 0.2181 | No |
| 34 | Ift88 | 6108 | 0.149 | 0.2155 | No |
| 35 | Oaz3 | 6474 | 0.133 | 0.2023 | No |
| 36 | Snap91 | 6588 | 0.128 | 0.2017 | No |
| 37 | Zc3h14 | 6641 | 0.124 | 0.2041 | No |
| 38 | Map7 | 6751 | 0.119 | 0.2034 | No |
| 39 | Gfi1 | 7070 | 0.104 | 0.1914 | No |
| 40 | Gstm5 | 7094 | 0.103 | 0.1944 | No |
| 41 | Rad17 | 7472 | 0.086 | 0.1787 | No |
| 42 | Strbp | 7490 | 0.085 | 0.1812 | No |
| 43 | Psmg1 | 7730 | 0.074 | 0.1721 | No |
| 44 | Nf2 | 7922 | 0.066 | 0.1651 | No |
| 45 | Ip6k1 | 8010 | 0.063 | 0.1632 | No |
| 46 | Ezh2 | 8072 | 0.059 | 0.1625 | No |
| 47 | Tcp11 | 8165 | 0.054 | 0.1600 | No |
| 48 | Pomc | 8353 | 0.046 | 0.1524 | No |
| 49 | Mtor | 8358 | 0.046 | 0.1540 | No |
| 50 | Chfr | 8581 | 0.036 | 0.1442 | No |
| 51 | Stam2 | 9557 | -0.000 | 0.0946 | No |
| 52 | Tsn | 9826 | -0.011 | 0.0815 | No |
| 53 | Parp2 | 9909 | -0.015 | 0.0779 | No |
| 54 | Phf7 | 9952 | -0.017 | 0.0765 | No |
| 55 | Pias2 | 10116 | -0.023 | 0.0691 | No |
| 56 | Csnk2a2 | 10407 | -0.036 | 0.0558 | No |
| 57 | Septin4 | 10446 | -0.038 | 0.0554 | No |
| 58 | Tle4 | 10688 | -0.047 | 0.0451 | No |
| 59 | Slc12a2 | 10714 | -0.048 | 0.0458 | No |
| 60 | Clpb | 11191 | -0.067 | 0.0243 | No |
| 61 | Nphp1 | 11388 | -0.075 | 0.0174 | No |
| 62 | She | 11989 | -0.101 | -0.0091 | No |
| 63 | Pacrg | 12121 | -0.107 | -0.0114 | No |
| 64 | Tulp2 | 12142 | -0.108 | -0.0080 | No |
| 65 | Mast2 | 12500 | -0.123 | -0.0212 | No |
| 66 | Zpbp | 12617 | -0.129 | -0.0219 | No |
| 67 | Spata6 | 12635 | -0.129 | -0.0175 | No |
| 68 | Braf | 12713 | -0.133 | -0.0161 | No |
| 69 | Lpin1 | 12985 | -0.145 | -0.0240 | No |
| 70 | Tnni3 | 13294 | -0.157 | -0.0333 | No |
| 71 | Sirt1 | 14066 | -0.192 | -0.0648 | No |
| 72 | Slc2a5 | 14075 | -0.192 | -0.0574 | No |
| 73 | Mlf1 | 14116 | -0.193 | -0.0516 | No |
| 74 | Grm8 | 14250 | -0.200 | -0.0503 | No |
| 75 | Mllt10 | 14506 | -0.212 | -0.0547 | No |
| 76 | Dcc | 14837 | -0.229 | -0.0622 | No |
| 77 | Cct6b | 15009 | -0.237 | -0.0614 | No |
| 78 | Phkg2 | 15013 | -0.237 | -0.0519 | No |
| 79 | Dnajb8 | 15110 | -0.242 | -0.0470 | No |
| 80 | Gmcl1 | 15407 | -0.258 | -0.0517 | No |
| 81 | Adam2 | 15489 | -0.261 | -0.0452 | No |
| 82 | Hspa1l | 15607 | -0.267 | -0.0404 | No |
| 83 | Pcsk4 | 15981 | -0.289 | -0.0477 | No |
| 84 | Cnih2 | 16146 | -0.299 | -0.0439 | No |
| 85 | Cftr | 16239 | -0.304 | -0.0363 | No |
| 86 | Gpr182 | 16617 | -0.328 | -0.0422 | No |
| 87 | Il12rb2 | 16742 | -0.337 | -0.0349 | No |
| 88 | Taldo1 | 16828 | -0.344 | -0.0254 | No |
| 89 | Adad1 | 17018 | -0.358 | -0.0205 | No |
| 90 | Cdkn3 | 18177 | -0.455 | -0.0610 | No |
| 91 | Acrbp | 18207 | -0.459 | -0.0439 | No |
| 92 | Ddx4 | 18325 | -0.474 | -0.0307 | No |
| 93 | Crisp2 | 18448 | -0.489 | -0.0171 | No |
| 94 | Ybx2 | 18676 | -0.523 | -0.0076 | No |
| 95 | Scg5 | 19444 | -0.707 | -0.0180 | No |
| 96 | Camk4 | 19638 | -0.856 | 0.0068 | No |