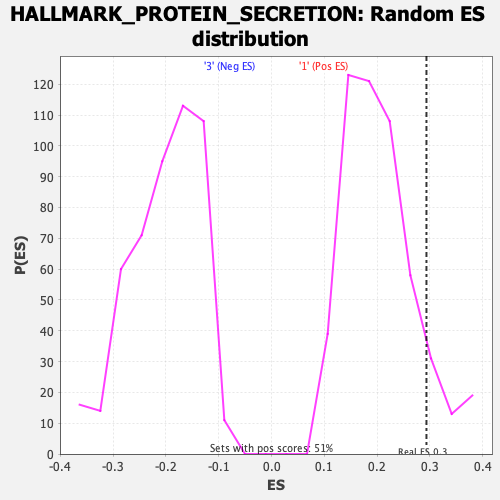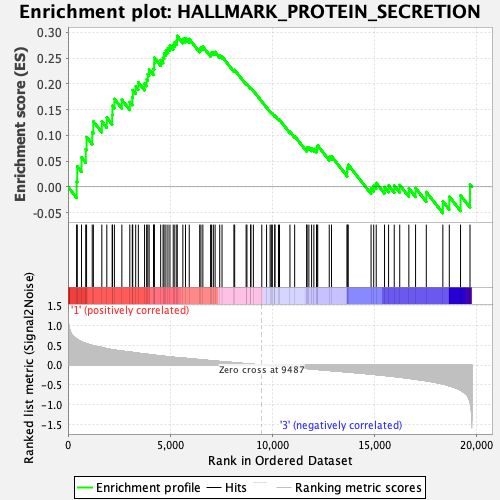
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group4.CMP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.29333726 |
| Normalized Enrichment Score (NES) | 1.4305962 |
| Nominal p-value | 0.1015625 |
| FDR q-value | 0.19506496 |
| FWER p-Value | 0.512 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cltc | 421 | 0.657 | 0.0103 | Yes |
| 2 | Rps6ka3 | 454 | 0.645 | 0.0397 | Yes |
| 3 | Dst | 662 | 0.583 | 0.0573 | Yes |
| 4 | Anp32e | 868 | 0.542 | 0.0730 | Yes |
| 5 | Gla | 909 | 0.536 | 0.0968 | Yes |
| 6 | Gosr2 | 1186 | 0.491 | 0.1064 | Yes |
| 7 | Sec24d | 1242 | 0.485 | 0.1270 | Yes |
| 8 | Adam10 | 1656 | 0.438 | 0.1271 | Yes |
| 9 | Arf1 | 1900 | 0.411 | 0.1345 | Yes |
| 10 | Ykt6 | 2164 | 0.383 | 0.1397 | Yes |
| 11 | Arcn1 | 2176 | 0.383 | 0.1575 | Yes |
| 12 | Atp1a1 | 2272 | 0.375 | 0.1708 | Yes |
| 13 | Copb2 | 2637 | 0.348 | 0.1691 | Yes |
| 14 | Lman1 | 3026 | 0.322 | 0.1649 | Yes |
| 15 | Uso1 | 3148 | 0.316 | 0.1740 | Yes |
| 16 | Clcn3 | 3170 | 0.314 | 0.1880 | Yes |
| 17 | Stx16 | 3314 | 0.301 | 0.1953 | Yes |
| 18 | Vamp3 | 3444 | 0.295 | 0.2029 | Yes |
| 19 | Atp6v1h | 3748 | 0.277 | 0.2009 | Yes |
| 20 | Napg | 3850 | 0.271 | 0.2088 | Yes |
| 21 | Copb1 | 3907 | 0.267 | 0.2188 | Yes |
| 22 | Arfgef2 | 3972 | 0.262 | 0.2282 | Yes |
| 23 | Vps4b | 4186 | 0.248 | 0.2294 | Yes |
| 24 | Ap2m1 | 4220 | 0.245 | 0.2395 | Yes |
| 25 | Tsg101 | 4226 | 0.245 | 0.2511 | Yes |
| 26 | Tmed10 | 4537 | 0.227 | 0.2462 | Yes |
| 27 | Cln5 | 4655 | 0.222 | 0.2510 | Yes |
| 28 | Rab14 | 4704 | 0.218 | 0.2591 | Yes |
| 29 | Zw10 | 4794 | 0.212 | 0.2647 | Yes |
| 30 | Cog2 | 4894 | 0.208 | 0.2697 | Yes |
| 31 | Tmx1 | 4992 | 0.201 | 0.2745 | Yes |
| 32 | Mon2 | 5152 | 0.194 | 0.2758 | Yes |
| 33 | Scrn1 | 5230 | 0.189 | 0.2810 | Yes |
| 34 | Scamp1 | 5332 | 0.187 | 0.2848 | Yes |
| 35 | Snx2 | 5342 | 0.186 | 0.2933 | Yes |
| 36 | Golga4 | 5623 | 0.171 | 0.2874 | No |
| 37 | Arfip1 | 5752 | 0.167 | 0.2889 | No |
| 38 | Sh3gl2 | 5936 | 0.159 | 0.2873 | No |
| 39 | Tpd52 | 6448 | 0.134 | 0.2678 | No |
| 40 | Pam | 6509 | 0.131 | 0.2711 | No |
| 41 | Ctsc | 6603 | 0.127 | 0.2724 | No |
| 42 | Arfgap3 | 6981 | 0.108 | 0.2585 | No |
| 43 | M6pr | 7030 | 0.105 | 0.2611 | No |
| 44 | Clta | 7116 | 0.102 | 0.2617 | No |
| 45 | Lamp2 | 7203 | 0.098 | 0.2620 | No |
| 46 | Mapk1 | 7428 | 0.088 | 0.2549 | No |
| 47 | Krt18 | 7541 | 0.082 | 0.2532 | No |
| 48 | Rab9 | 8123 | 0.056 | 0.2263 | No |
| 49 | Sec31a | 8161 | 0.055 | 0.2271 | No |
| 50 | Dnm1l | 8721 | 0.031 | 0.2002 | No |
| 51 | Arfgef1 | 8761 | 0.029 | 0.1996 | No |
| 52 | Dop1a | 8944 | 0.021 | 0.1914 | No |
| 53 | Kif1b | 8954 | 0.021 | 0.1919 | No |
| 54 | Ocrl | 9076 | 0.016 | 0.1866 | No |
| 55 | Egfr | 9488 | 0.000 | 0.1657 | No |
| 56 | Ap2b1 | 9722 | -0.007 | 0.1542 | No |
| 57 | Sec22b | 9896 | -0.014 | 0.1461 | No |
| 58 | Ap3b1 | 9955 | -0.017 | 0.1440 | No |
| 59 | Cope | 10005 | -0.019 | 0.1424 | No |
| 60 | Igf2r | 10123 | -0.024 | 0.1376 | No |
| 61 | Gbf1 | 10132 | -0.024 | 0.1384 | No |
| 62 | Galc | 10305 | -0.032 | 0.1312 | No |
| 63 | Stx7 | 10356 | -0.034 | 0.1303 | No |
| 64 | Stx12 | 10867 | -0.055 | 0.1070 | No |
| 65 | Ap1g1 | 11097 | -0.065 | 0.0985 | No |
| 66 | Vps45 | 11684 | -0.088 | 0.0729 | No |
| 67 | Rab22a | 11705 | -0.088 | 0.0762 | No |
| 68 | Rab2a | 11793 | -0.092 | 0.0762 | No |
| 69 | Atp7a | 11926 | -0.098 | 0.0742 | No |
| 70 | Cav2 | 12038 | -0.103 | 0.0736 | No |
| 71 | Ppt1 | 12168 | -0.109 | 0.0723 | No |
| 72 | Vamp4 | 12193 | -0.110 | 0.0764 | No |
| 73 | Snap23 | 12227 | -0.112 | 0.0801 | No |
| 74 | Rer1 | 12791 | -0.137 | 0.0580 | No |
| 75 | Ica1 | 12902 | -0.142 | 0.0593 | No |
| 76 | Sod1 | 13666 | -0.173 | 0.0289 | No |
| 77 | Ap2s1 | 13670 | -0.174 | 0.0371 | No |
| 78 | Ap3s1 | 13721 | -0.176 | 0.0430 | No |
| 79 | Sgms1 | 14839 | -0.229 | -0.0027 | No |
| 80 | Stam | 14968 | -0.235 | 0.0021 | No |
| 81 | Rab5a | 15091 | -0.241 | 0.0076 | No |
| 82 | Scamp3 | 15497 | -0.262 | -0.0004 | No |
| 83 | Napa | 15696 | -0.272 | 0.0027 | No |
| 84 | Yipf6 | 15973 | -0.288 | 0.0025 | No |
| 85 | Tspan8 | 16242 | -0.304 | 0.0036 | No |
| 86 | Cd63 | 16688 | -0.333 | -0.0030 | No |
| 87 | Ergic3 | 17017 | -0.358 | -0.0024 | No |
| 88 | Gnas | 17543 | -0.395 | -0.0101 | No |
| 89 | Tom1l1 | 18353 | -0.476 | -0.0282 | No |
| 90 | Abca1 | 18669 | -0.522 | -0.0190 | No |
| 91 | Bet1 | 19219 | -0.629 | -0.0166 | No |
| 92 | Bnip3 | 19682 | -0.925 | 0.0045 | No |