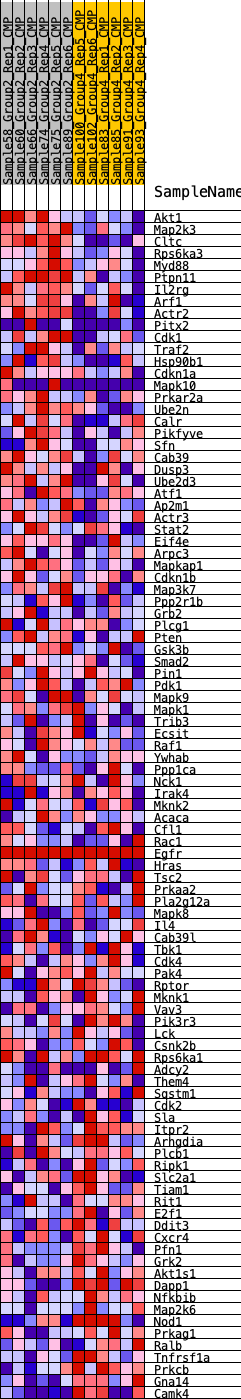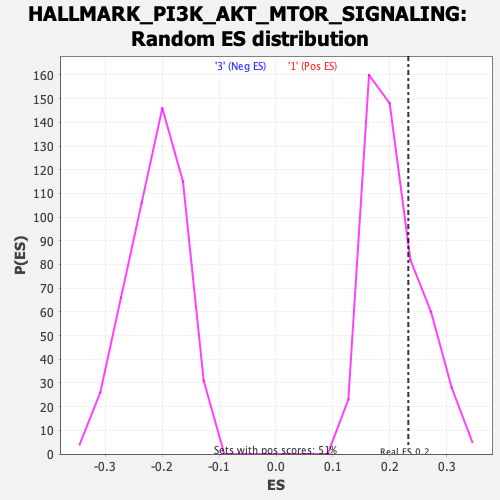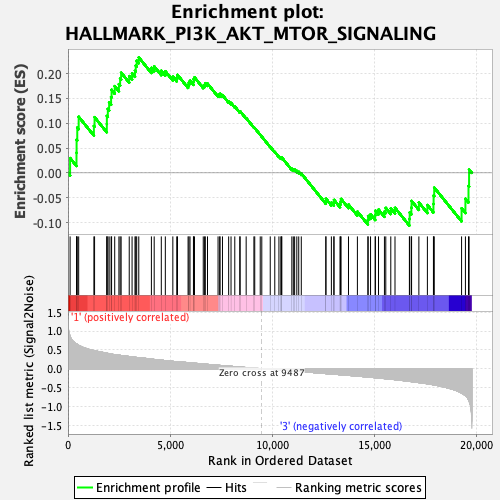
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group4.CMP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.2323522 |
| Normalized Enrichment Score (NES) | 1.1284926 |
| Nominal p-value | 0.24901186 |
| FDR q-value | 0.46246007 |
| FWER p-Value | 0.905 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Akt1 | 103 | 0.864 | 0.0297 | Yes |
| 2 | Map2k3 | 416 | 0.659 | 0.0405 | Yes |
| 3 | Cltc | 421 | 0.657 | 0.0668 | Yes |
| 4 | Rps6ka3 | 454 | 0.645 | 0.0913 | Yes |
| 5 | Myd88 | 523 | 0.620 | 0.1128 | Yes |
| 6 | Ptpn11 | 1268 | 0.480 | 0.0944 | Yes |
| 7 | Il2rg | 1301 | 0.476 | 0.1120 | Yes |
| 8 | Arf1 | 1900 | 0.411 | 0.0982 | Yes |
| 9 | Actr2 | 1903 | 0.410 | 0.1147 | Yes |
| 10 | Pitx2 | 1950 | 0.406 | 0.1288 | Yes |
| 11 | Cdk1 | 2014 | 0.400 | 0.1418 | Yes |
| 12 | Traf2 | 2110 | 0.390 | 0.1527 | Yes |
| 13 | Hsp90b1 | 2134 | 0.387 | 0.1671 | Yes |
| 14 | Cdkn1a | 2287 | 0.374 | 0.1745 | Yes |
| 15 | Mapk10 | 2493 | 0.358 | 0.1786 | Yes |
| 16 | Prkar2a | 2552 | 0.355 | 0.1900 | Yes |
| 17 | Ube2n | 2601 | 0.351 | 0.2017 | Yes |
| 18 | Calr | 2998 | 0.324 | 0.1947 | Yes |
| 19 | Pikfyve | 3137 | 0.317 | 0.2005 | Yes |
| 20 | Sfn | 3280 | 0.304 | 0.2055 | Yes |
| 21 | Cab39 | 3317 | 0.301 | 0.2159 | Yes |
| 22 | Dusp3 | 3363 | 0.298 | 0.2256 | Yes |
| 23 | Ube2d3 | 3465 | 0.293 | 0.2324 | Yes |
| 24 | Atf1 | 4083 | 0.255 | 0.2113 | No |
| 25 | Ap2m1 | 4220 | 0.245 | 0.2143 | No |
| 26 | Actr3 | 4568 | 0.225 | 0.2057 | No |
| 27 | Stat2 | 4765 | 0.214 | 0.2044 | No |
| 28 | Eif4e | 5133 | 0.195 | 0.1936 | No |
| 29 | Arpc3 | 5324 | 0.187 | 0.1915 | No |
| 30 | Mapkap1 | 5361 | 0.185 | 0.1972 | No |
| 31 | Cdkn1b | 5879 | 0.162 | 0.1775 | No |
| 32 | Map3k7 | 5921 | 0.160 | 0.1818 | No |
| 33 | Ppp2r1b | 5974 | 0.156 | 0.1855 | No |
| 34 | Grb2 | 6142 | 0.147 | 0.1830 | No |
| 35 | Plcg1 | 6149 | 0.147 | 0.1886 | No |
| 36 | Pten | 6193 | 0.144 | 0.1922 | No |
| 37 | Gsk3b | 6622 | 0.126 | 0.1756 | No |
| 38 | Smad2 | 6688 | 0.122 | 0.1772 | No |
| 39 | Pin1 | 6722 | 0.120 | 0.1804 | No |
| 40 | Pdk1 | 6830 | 0.115 | 0.1796 | No |
| 41 | Mapk9 | 7347 | 0.091 | 0.1570 | No |
| 42 | Mapk1 | 7428 | 0.088 | 0.1565 | No |
| 43 | Trib3 | 7443 | 0.087 | 0.1593 | No |
| 44 | Ecsit | 7563 | 0.081 | 0.1565 | No |
| 45 | Raf1 | 7866 | 0.068 | 0.1440 | No |
| 46 | Ywhab | 7982 | 0.064 | 0.1407 | No |
| 47 | Ppp1ca | 8166 | 0.054 | 0.1336 | No |
| 48 | Nck1 | 8410 | 0.044 | 0.1230 | No |
| 49 | Irak4 | 8428 | 0.043 | 0.1239 | No |
| 50 | Mknk2 | 8723 | 0.031 | 0.1102 | No |
| 51 | Acaca | 9102 | 0.015 | 0.0916 | No |
| 52 | Cfl1 | 9143 | 0.014 | 0.0901 | No |
| 53 | Rac1 | 9412 | 0.003 | 0.0766 | No |
| 54 | Egfr | 9488 | 0.000 | 0.0728 | No |
| 55 | Hras | 9902 | -0.014 | 0.0524 | No |
| 56 | Tsc2 | 10124 | -0.024 | 0.0421 | No |
| 57 | Prkaa2 | 10327 | -0.032 | 0.0332 | No |
| 58 | Pla2g12a | 10420 | -0.037 | 0.0300 | No |
| 59 | Mapk8 | 10460 | -0.038 | 0.0295 | No |
| 60 | Il4 | 10464 | -0.039 | 0.0309 | No |
| 61 | Cab39l | 10960 | -0.059 | 0.0082 | No |
| 62 | Tbk1 | 11032 | -0.063 | 0.0071 | No |
| 63 | Cdk4 | 11096 | -0.065 | 0.0065 | No |
| 64 | Pak4 | 11203 | -0.067 | 0.0039 | No |
| 65 | Rptor | 11297 | -0.072 | 0.0020 | No |
| 66 | Mknk1 | 11422 | -0.076 | -0.0012 | No |
| 67 | Vav3 | 12623 | -0.129 | -0.0570 | No |
| 68 | Pik3r3 | 12629 | -0.129 | -0.0520 | No |
| 69 | Lck | 12895 | -0.142 | -0.0598 | No |
| 70 | Csnk2b | 13016 | -0.146 | -0.0600 | No |
| 71 | Rps6ka1 | 13025 | -0.146 | -0.0545 | No |
| 72 | Adcy2 | 13318 | -0.158 | -0.0629 | No |
| 73 | Them4 | 13344 | -0.159 | -0.0577 | No |
| 74 | Sqstm1 | 13368 | -0.160 | -0.0524 | No |
| 75 | Cdk2 | 13733 | -0.177 | -0.0638 | No |
| 76 | Sla | 14169 | -0.196 | -0.0780 | No |
| 77 | Itpr2 | 14682 | -0.221 | -0.0951 | No |
| 78 | Arhgdia | 14696 | -0.222 | -0.0868 | No |
| 79 | Plcb1 | 14813 | -0.228 | -0.0835 | No |
| 80 | Ripk1 | 15040 | -0.238 | -0.0853 | No |
| 81 | Slc2a1 | 15050 | -0.239 | -0.0761 | No |
| 82 | Tiam1 | 15203 | -0.247 | -0.0738 | No |
| 83 | Rit1 | 15493 | -0.262 | -0.0779 | No |
| 84 | E2f1 | 15559 | -0.265 | -0.0705 | No |
| 85 | Ddit3 | 15804 | -0.279 | -0.0717 | No |
| 86 | Cxcr4 | 16010 | -0.291 | -0.0703 | No |
| 87 | Pfn1 | 16715 | -0.335 | -0.0926 | No |
| 88 | Grk2 | 16730 | -0.336 | -0.0797 | No |
| 89 | Akt1s1 | 16811 | -0.343 | -0.0699 | No |
| 90 | Dapp1 | 16822 | -0.343 | -0.0565 | No |
| 91 | Nfkbib | 17179 | -0.368 | -0.0597 | No |
| 92 | Map2k6 | 17596 | -0.399 | -0.0648 | No |
| 93 | Nod1 | 17892 | -0.428 | -0.0625 | No |
| 94 | Prkag1 | 17901 | -0.428 | -0.0456 | No |
| 95 | Ralb | 17931 | -0.431 | -0.0296 | No |
| 96 | Tnfrsf1a | 19270 | -0.649 | -0.0714 | No |
| 97 | Prkcb | 19458 | -0.714 | -0.0520 | No |
| 98 | Gna14 | 19609 | -0.822 | -0.0264 | No |
| 99 | Camk4 | 19638 | -0.856 | 0.0068 | No |