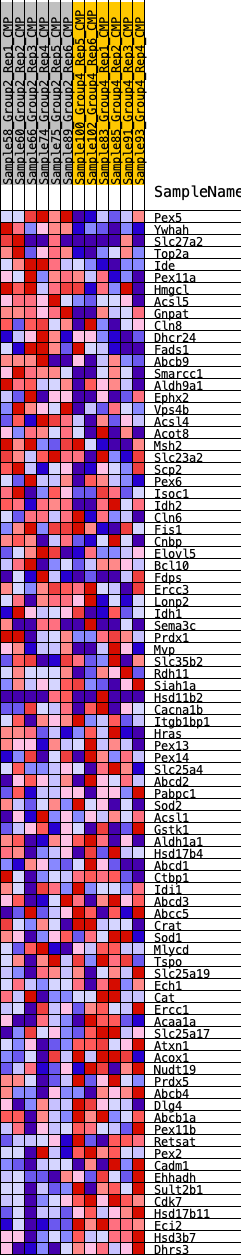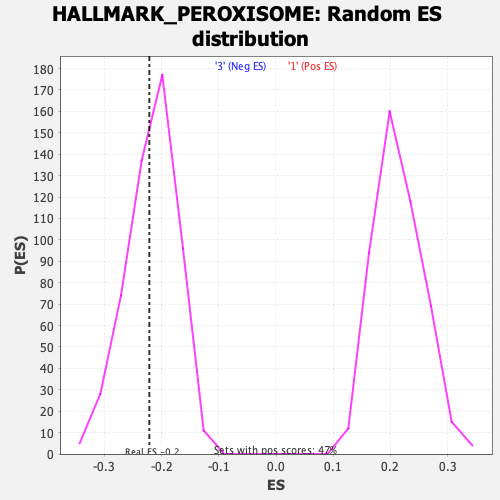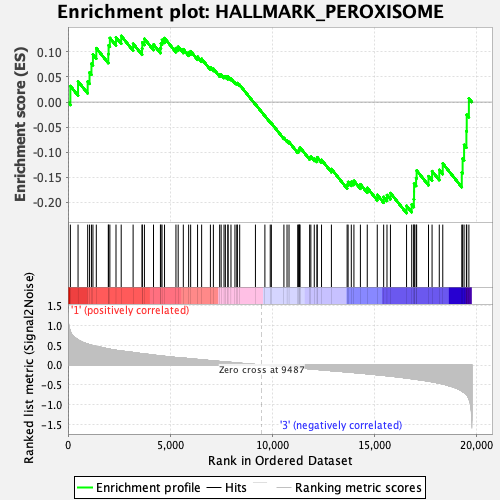
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group4.CMP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.22133872 |
| Normalized Enrichment Score (NES) | -1.0210104 |
| Nominal p-value | 0.4034091 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.979 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex5 | 116 | 0.844 | 0.0320 | No |
| 2 | Ywhah | 492 | 0.630 | 0.0412 | No |
| 3 | Slc27a2 | 964 | 0.527 | 0.0409 | No |
| 4 | Top2a | 1055 | 0.507 | 0.0591 | No |
| 5 | Ide | 1148 | 0.496 | 0.0767 | No |
| 6 | Pex11a | 1221 | 0.486 | 0.0948 | No |
| 7 | Hmgcl | 1386 | 0.466 | 0.1074 | No |
| 8 | Acsl5 | 1973 | 0.403 | 0.0957 | No |
| 9 | Gnpat | 1988 | 0.402 | 0.1130 | No |
| 10 | Cln8 | 2050 | 0.397 | 0.1277 | No |
| 11 | Dhcr24 | 2350 | 0.368 | 0.1291 | No |
| 12 | Fads1 | 2605 | 0.350 | 0.1319 | No |
| 13 | Abcb9 | 3188 | 0.312 | 0.1163 | No |
| 14 | Smarcc1 | 3626 | 0.283 | 0.1068 | No |
| 15 | Aldh9a1 | 3638 | 0.282 | 0.1189 | No |
| 16 | Ephx2 | 3742 | 0.278 | 0.1261 | No |
| 17 | Vps4b | 4186 | 0.248 | 0.1147 | No |
| 18 | Acsl4 | 4525 | 0.227 | 0.1077 | No |
| 19 | Acot8 | 4549 | 0.226 | 0.1167 | No |
| 20 | Msh2 | 4604 | 0.223 | 0.1240 | No |
| 21 | Slc23a2 | 4726 | 0.217 | 0.1275 | No |
| 22 | Scp2 | 5285 | 0.189 | 0.1076 | No |
| 23 | Pex6 | 5394 | 0.184 | 0.1104 | No |
| 24 | Isoc1 | 5646 | 0.170 | 0.1053 | No |
| 25 | Idh2 | 5907 | 0.160 | 0.0993 | No |
| 26 | Cln6 | 6005 | 0.155 | 0.1013 | No |
| 27 | Fis1 | 6343 | 0.138 | 0.0904 | No |
| 28 | Cnbp | 6543 | 0.130 | 0.0861 | No |
| 29 | Elovl5 | 6972 | 0.108 | 0.0692 | No |
| 30 | Bcl10 | 7118 | 0.102 | 0.0664 | No |
| 31 | Fdps | 7426 | 0.088 | 0.0548 | No |
| 32 | Ercc3 | 7498 | 0.085 | 0.0549 | No |
| 33 | Lonp2 | 7639 | 0.078 | 0.0513 | No |
| 34 | Idh1 | 7705 | 0.075 | 0.0514 | No |
| 35 | Sema3c | 7829 | 0.070 | 0.0483 | No |
| 36 | Prdx1 | 7835 | 0.070 | 0.0512 | No |
| 37 | Mvp | 7979 | 0.064 | 0.0468 | No |
| 38 | Slc35b2 | 8174 | 0.054 | 0.0393 | No |
| 39 | Rdh11 | 8265 | 0.050 | 0.0370 | No |
| 40 | Siah1a | 8293 | 0.050 | 0.0379 | No |
| 41 | Hsd11b2 | 8406 | 0.044 | 0.0342 | No |
| 42 | Cacna1b | 9177 | 0.012 | -0.0044 | No |
| 43 | Itgb1bp1 | 9640 | -0.004 | -0.0277 | No |
| 44 | Hras | 9902 | -0.014 | -0.0403 | No |
| 45 | Pex13 | 9957 | -0.017 | -0.0423 | No |
| 46 | Pex14 | 10571 | -0.043 | -0.0715 | No |
| 47 | Slc25a4 | 10724 | -0.049 | -0.0770 | No |
| 48 | Abcd2 | 10821 | -0.053 | -0.0795 | No |
| 49 | Pabpc1 | 11247 | -0.069 | -0.0980 | No |
| 50 | Sod2 | 11292 | -0.071 | -0.0970 | No |
| 51 | Acsl1 | 11312 | -0.072 | -0.0948 | No |
| 52 | Gstk1 | 11333 | -0.073 | -0.0925 | No |
| 53 | Aldh1a1 | 11362 | -0.074 | -0.0906 | No |
| 54 | Hsd17b4 | 11831 | -0.093 | -0.1102 | No |
| 55 | Abcd1 | 11890 | -0.096 | -0.1088 | No |
| 56 | Ctbp1 | 12058 | -0.104 | -0.1126 | No |
| 57 | Idi1 | 12191 | -0.110 | -0.1144 | No |
| 58 | Abcd3 | 12202 | -0.111 | -0.1099 | No |
| 59 | Abcc5 | 12413 | -0.120 | -0.1152 | No |
| 60 | Crat | 12897 | -0.142 | -0.1334 | No |
| 61 | Sod1 | 13666 | -0.173 | -0.1646 | No |
| 62 | Mlycd | 13714 | -0.176 | -0.1591 | No |
| 63 | Tspo | 13871 | -0.183 | -0.1589 | No |
| 64 | Slc25a19 | 13999 | -0.189 | -0.1568 | No |
| 65 | Ech1 | 14316 | -0.204 | -0.1638 | No |
| 66 | Cat | 14652 | -0.220 | -0.1709 | No |
| 67 | Ercc1 | 15142 | -0.244 | -0.1848 | No |
| 68 | Acaa1a | 15455 | -0.260 | -0.1890 | No |
| 69 | Slc25a17 | 15618 | -0.268 | -0.1852 | No |
| 70 | Atxn1 | 15787 | -0.278 | -0.1813 | No |
| 71 | Acox1 | 16576 | -0.325 | -0.2068 | Yes |
| 72 | Nudt19 | 16829 | -0.344 | -0.2041 | Yes |
| 73 | Prdx5 | 16933 | -0.352 | -0.1936 | Yes |
| 74 | Abcb4 | 16944 | -0.352 | -0.1783 | Yes |
| 75 | Dlg4 | 16945 | -0.352 | -0.1625 | Yes |
| 76 | Abcb1a | 17043 | -0.359 | -0.1513 | Yes |
| 77 | Pex11b | 17070 | -0.361 | -0.1364 | Yes |
| 78 | Retsat | 17651 | -0.403 | -0.1477 | Yes |
| 79 | Pex2 | 17825 | -0.420 | -0.1377 | Yes |
| 80 | Cadm1 | 18178 | -0.455 | -0.1351 | Yes |
| 81 | Ehhadh | 18349 | -0.476 | -0.1224 | Yes |
| 82 | Sult2b1 | 19281 | -0.653 | -0.1404 | Yes |
| 83 | Cdk7 | 19321 | -0.665 | -0.1125 | Yes |
| 84 | Hsd17b11 | 19393 | -0.688 | -0.0853 | Yes |
| 85 | Eci2 | 19506 | -0.736 | -0.0579 | Yes |
| 86 | Hsd3b7 | 19523 | -0.746 | -0.0253 | Yes |
| 87 | Dhrs3 | 19628 | -0.844 | 0.0073 | Yes |