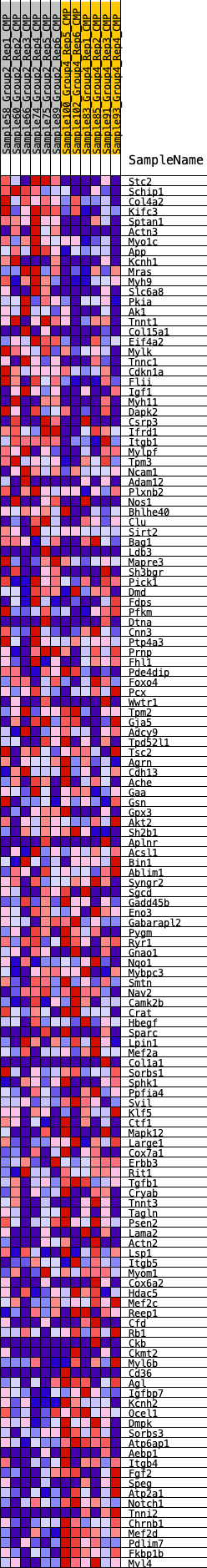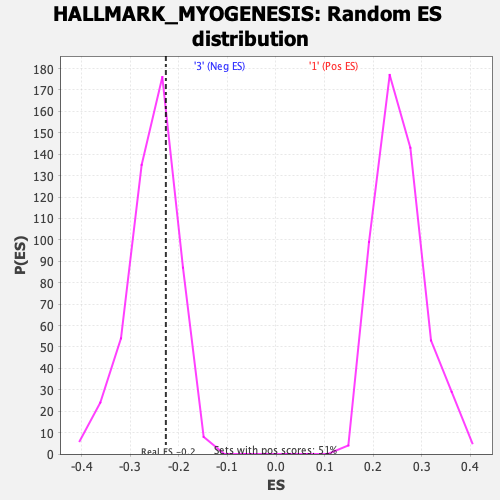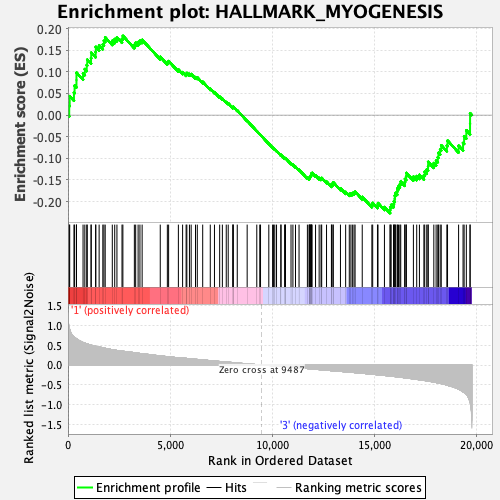
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group4.CMP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | -0.2266292 |
| Normalized Enrichment Score (NES) | -0.88800967 |
| Nominal p-value | 0.6877551 |
| FDR q-value | 0.9371855 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Stc2 | 60 | 0.944 | 0.0216 | No |
| 2 | Schip1 | 83 | 0.893 | 0.0438 | No |
| 3 | Col4a2 | 292 | 0.708 | 0.0517 | No |
| 4 | Kifc3 | 323 | 0.694 | 0.0683 | No |
| 5 | Sptan1 | 414 | 0.659 | 0.0809 | No |
| 6 | Actn3 | 415 | 0.659 | 0.0981 | No |
| 7 | Myo1c | 737 | 0.565 | 0.0965 | No |
| 8 | App | 824 | 0.548 | 0.1064 | No |
| 9 | Kcnh1 | 918 | 0.534 | 0.1156 | No |
| 10 | Mras | 943 | 0.530 | 0.1282 | No |
| 11 | Myh9 | 1128 | 0.498 | 0.1319 | No |
| 12 | Slc6a8 | 1137 | 0.497 | 0.1445 | No |
| 13 | Pkia | 1353 | 0.470 | 0.1458 | No |
| 14 | Ak1 | 1361 | 0.469 | 0.1577 | No |
| 15 | Tnnt1 | 1524 | 0.453 | 0.1612 | No |
| 16 | Col15a1 | 1705 | 0.433 | 0.1634 | No |
| 17 | Eif4a2 | 1759 | 0.427 | 0.1718 | No |
| 18 | Mylk | 1821 | 0.419 | 0.1796 | No |
| 19 | Tnnc1 | 2169 | 0.383 | 0.1720 | No |
| 20 | Cdkn1a | 2287 | 0.374 | 0.1758 | No |
| 21 | Flii | 2396 | 0.365 | 0.1798 | No |
| 22 | Igf1 | 2644 | 0.347 | 0.1763 | No |
| 23 | Myh11 | 2687 | 0.344 | 0.1831 | No |
| 24 | Dapk2 | 3249 | 0.308 | 0.1626 | No |
| 25 | Csrp3 | 3313 | 0.302 | 0.1673 | No |
| 26 | Ifrd1 | 3433 | 0.296 | 0.1689 | No |
| 27 | Itgb1 | 3522 | 0.289 | 0.1720 | No |
| 28 | Mylpf | 3630 | 0.282 | 0.1739 | No |
| 29 | Tpm3 | 4517 | 0.228 | 0.1347 | No |
| 30 | Ncam1 | 4864 | 0.209 | 0.1226 | No |
| 31 | Adam12 | 4926 | 0.206 | 0.1248 | No |
| 32 | Plxnb2 | 5404 | 0.183 | 0.1053 | No |
| 33 | Nos1 | 5611 | 0.172 | 0.0993 | No |
| 34 | Bhlhe40 | 5783 | 0.166 | 0.0949 | No |
| 35 | Clu | 5819 | 0.164 | 0.0974 | No |
| 36 | Sirt2 | 5946 | 0.158 | 0.0951 | No |
| 37 | Bag1 | 6030 | 0.153 | 0.0949 | No |
| 38 | Ldb3 | 6243 | 0.141 | 0.0878 | No |
| 39 | Mapre3 | 6330 | 0.139 | 0.0870 | No |
| 40 | Sh3bgr | 6595 | 0.128 | 0.0769 | No |
| 41 | Pick1 | 6966 | 0.108 | 0.0609 | No |
| 42 | Dmd | 7171 | 0.099 | 0.0531 | No |
| 43 | Fdps | 7426 | 0.088 | 0.0425 | No |
| 44 | Pfkm | 7558 | 0.082 | 0.0379 | No |
| 45 | Dtna | 7748 | 0.074 | 0.0302 | No |
| 46 | Cnn3 | 7844 | 0.070 | 0.0272 | No |
| 47 | Ptp4a3 | 8071 | 0.060 | 0.0172 | No |
| 48 | Prnp | 8084 | 0.058 | 0.0181 | No |
| 49 | Fhl1 | 8087 | 0.058 | 0.0196 | No |
| 50 | Pde4dip | 8287 | 0.050 | 0.0107 | No |
| 51 | Foxo4 | 8772 | 0.029 | -0.0132 | No |
| 52 | Pcx | 9243 | 0.010 | -0.0369 | No |
| 53 | Wwtr1 | 9389 | 0.003 | -0.0442 | No |
| 54 | Tpm2 | 9426 | 0.002 | -0.0459 | No |
| 55 | Gja5 | 9832 | -0.011 | -0.0663 | No |
| 56 | Adcy9 | 10017 | -0.020 | -0.0751 | No |
| 57 | Tpd52l1 | 10065 | -0.022 | -0.0769 | No |
| 58 | Tsc2 | 10124 | -0.024 | -0.0793 | No |
| 59 | Agrn | 10210 | -0.027 | -0.0829 | No |
| 60 | Cdh13 | 10424 | -0.037 | -0.0928 | No |
| 61 | Ache | 10427 | -0.037 | -0.0919 | No |
| 62 | Gaa | 10606 | -0.044 | -0.0998 | No |
| 63 | Gsn | 10633 | -0.045 | -0.1000 | No |
| 64 | Gpx3 | 10647 | -0.046 | -0.0994 | No |
| 65 | Akt2 | 10919 | -0.057 | -0.1117 | No |
| 66 | Sh2b1 | 11012 | -0.062 | -0.1148 | No |
| 67 | Aplnr | 11138 | -0.066 | -0.1194 | No |
| 68 | Acsl1 | 11312 | -0.072 | -0.1264 | No |
| 69 | Bin1 | 11721 | -0.089 | -0.1448 | No |
| 70 | Ablim1 | 11804 | -0.092 | -0.1466 | No |
| 71 | Syngr2 | 11815 | -0.093 | -0.1447 | No |
| 72 | Sgcd | 11838 | -0.094 | -0.1433 | No |
| 73 | Gadd45b | 11852 | -0.095 | -0.1415 | No |
| 74 | Eno3 | 11884 | -0.096 | -0.1406 | No |
| 75 | Gabarapl2 | 11898 | -0.097 | -0.1387 | No |
| 76 | Pygm | 11913 | -0.098 | -0.1369 | No |
| 77 | Ryr1 | 11920 | -0.098 | -0.1347 | No |
| 78 | Gnao1 | 11951 | -0.099 | -0.1336 | No |
| 79 | Nqo1 | 12120 | -0.107 | -0.1394 | No |
| 80 | Mybpc3 | 12296 | -0.115 | -0.1453 | No |
| 81 | Smtn | 12389 | -0.119 | -0.1469 | No |
| 82 | Nav2 | 12421 | -0.120 | -0.1453 | No |
| 83 | Camk2b | 12658 | -0.131 | -0.1539 | No |
| 84 | Crat | 12897 | -0.142 | -0.1623 | No |
| 85 | Hbegf | 12911 | -0.142 | -0.1593 | No |
| 86 | Sparc | 12976 | -0.145 | -0.1588 | No |
| 87 | Lpin1 | 12985 | -0.145 | -0.1554 | No |
| 88 | Mef2a | 13340 | -0.159 | -0.1693 | No |
| 89 | Col1a1 | 13594 | -0.172 | -0.1777 | No |
| 90 | Sorbs1 | 13772 | -0.178 | -0.1820 | No |
| 91 | Sphk1 | 13851 | -0.182 | -0.1813 | No |
| 92 | Ppfia4 | 13923 | -0.184 | -0.1801 | No |
| 93 | Svil | 14002 | -0.189 | -0.1791 | No |
| 94 | Klf5 | 14056 | -0.191 | -0.1768 | No |
| 95 | Ctf1 | 14406 | -0.207 | -0.1892 | No |
| 96 | Mapk12 | 14878 | -0.230 | -0.2071 | No |
| 97 | Large1 | 14918 | -0.232 | -0.2031 | No |
| 98 | Cox7a1 | 15152 | -0.244 | -0.2086 | No |
| 99 | Erbb3 | 15178 | -0.246 | -0.2034 | No |
| 100 | Rit1 | 15493 | -0.262 | -0.2126 | No |
| 101 | Tgfb1 | 15770 | -0.277 | -0.2194 | Yes |
| 102 | Cryab | 15776 | -0.277 | -0.2124 | Yes |
| 103 | Tnnt3 | 15822 | -0.279 | -0.2074 | Yes |
| 104 | Tagln | 15938 | -0.286 | -0.2058 | Yes |
| 105 | Psen2 | 15956 | -0.287 | -0.1992 | Yes |
| 106 | Lama2 | 15999 | -0.290 | -0.1938 | Yes |
| 107 | Actn2 | 16002 | -0.290 | -0.1863 | Yes |
| 108 | Lsp1 | 16028 | -0.292 | -0.1800 | Yes |
| 109 | Itgb5 | 16117 | -0.297 | -0.1767 | Yes |
| 110 | Myom1 | 16126 | -0.297 | -0.1693 | Yes |
| 111 | Cox6a2 | 16183 | -0.301 | -0.1643 | Yes |
| 112 | Hdac5 | 16249 | -0.305 | -0.1597 | Yes |
| 113 | Mef2c | 16291 | -0.308 | -0.1537 | Yes |
| 114 | Reep1 | 16485 | -0.320 | -0.1552 | Yes |
| 115 | Cfd | 16499 | -0.321 | -0.1475 | Yes |
| 116 | Rb1 | 16557 | -0.324 | -0.1419 | Yes |
| 117 | Ckb | 16564 | -0.325 | -0.1337 | Yes |
| 118 | Ckmt2 | 16911 | -0.351 | -0.1422 | Yes |
| 119 | Myl6b | 17073 | -0.361 | -0.1410 | Yes |
| 120 | Cd36 | 17206 | -0.371 | -0.1380 | Yes |
| 121 | Agl | 17425 | -0.386 | -0.1390 | Yes |
| 122 | Igfbp7 | 17465 | -0.389 | -0.1309 | Yes |
| 123 | Kcnh2 | 17565 | -0.396 | -0.1256 | Yes |
| 124 | Ocel1 | 17629 | -0.401 | -0.1183 | Yes |
| 125 | Dmpk | 17634 | -0.402 | -0.1080 | Yes |
| 126 | Sorbs3 | 17911 | -0.429 | -0.1109 | Yes |
| 127 | Atp6ap1 | 18023 | -0.440 | -0.1050 | Yes |
| 128 | Aebp1 | 18106 | -0.448 | -0.0975 | Yes |
| 129 | Itgb4 | 18136 | -0.450 | -0.0872 | Yes |
| 130 | Fgf2 | 18221 | -0.461 | -0.0794 | Yes |
| 131 | Speg | 18274 | -0.467 | -0.0699 | Yes |
| 132 | Atp2a1 | 18550 | -0.505 | -0.0707 | Yes |
| 133 | Notch1 | 18582 | -0.509 | -0.0590 | Yes |
| 134 | Tnni2 | 19124 | -0.605 | -0.0708 | Yes |
| 135 | Chrnb1 | 19342 | -0.673 | -0.0642 | Yes |
| 136 | Mef2d | 19394 | -0.688 | -0.0489 | Yes |
| 137 | Pdlim7 | 19503 | -0.735 | -0.0352 | Yes |
| 138 | Fkbp1b | 19685 | -0.932 | -0.0201 | Yes |
| 139 | Myl4 | 19687 | -0.935 | 0.0043 | Yes |