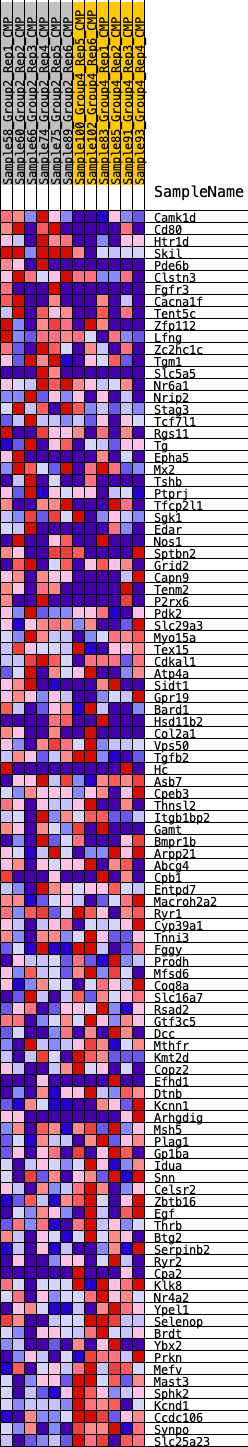
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group4.CMP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
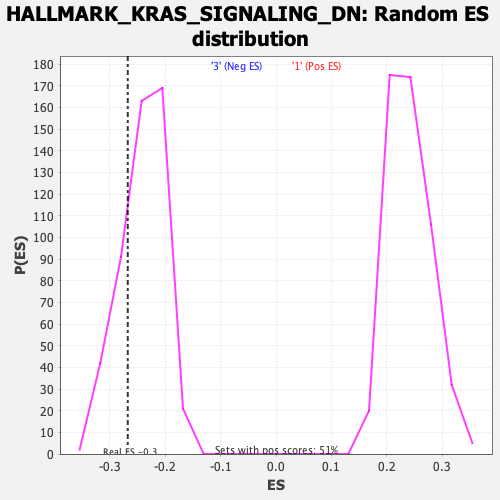
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.26731488 |
| Normalized Enrichment Score (NES) | -1.1163269 |
| Nominal p-value | 0.21106558 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.933 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Camk1d | 146 | 0.796 | 0.0211 | No |
| 2 | Cd80 | 433 | 0.652 | 0.0299 | No |
| 3 | Htr1d | 688 | 0.577 | 0.0377 | No |
| 4 | Skil | 750 | 0.562 | 0.0548 | No |
| 5 | Pde6b | 901 | 0.536 | 0.0663 | No |
| 6 | Clstn3 | 1124 | 0.499 | 0.0729 | No |
| 7 | Fgfr3 | 1609 | 0.444 | 0.0642 | No |
| 8 | Cacna1f | 2082 | 0.393 | 0.0543 | No |
| 9 | Tent5c | 2259 | 0.376 | 0.0589 | No |
| 10 | Zfp112 | 2375 | 0.366 | 0.0661 | No |
| 11 | Lfng | 2506 | 0.358 | 0.0723 | No |
| 12 | Zc2hc1c | 2521 | 0.357 | 0.0844 | No |
| 13 | Tgm1 | 2688 | 0.344 | 0.0883 | No |
| 14 | Slc5a5 | 2698 | 0.343 | 0.1001 | No |
| 15 | Nr6a1 | 2885 | 0.331 | 0.1025 | No |
| 16 | Nrip2 | 2981 | 0.325 | 0.1094 | No |
| 17 | Stag3 | 3135 | 0.317 | 0.1129 | No |
| 18 | Tcf7l1 | 3559 | 0.287 | 0.1017 | No |
| 19 | Rgs11 | 4121 | 0.252 | 0.0822 | No |
| 20 | Tg | 4237 | 0.244 | 0.0851 | No |
| 21 | Epha5 | 4444 | 0.232 | 0.0830 | No |
| 22 | Mx2 | 4509 | 0.228 | 0.0879 | No |
| 23 | Tshb | 4687 | 0.220 | 0.0868 | No |
| 24 | Ptprj | 4738 | 0.216 | 0.0919 | No |
| 25 | Tfcp2l1 | 5158 | 0.193 | 0.0776 | No |
| 26 | Sgk1 | 5200 | 0.191 | 0.0823 | No |
| 27 | Edar | 5537 | 0.176 | 0.0715 | No |
| 28 | Nos1 | 5611 | 0.172 | 0.0740 | No |
| 29 | Sptbn2 | 5893 | 0.161 | 0.0655 | No |
| 30 | Grid2 | 6075 | 0.151 | 0.0617 | No |
| 31 | Capn9 | 6094 | 0.150 | 0.0661 | No |
| 32 | Tenm2 | 6163 | 0.146 | 0.0679 | No |
| 33 | P2rx6 | 6508 | 0.132 | 0.0551 | No |
| 34 | Pdk2 | 6814 | 0.116 | 0.0438 | No |
| 35 | Slc29a3 | 6894 | 0.112 | 0.0438 | No |
| 36 | Myo15a | 7780 | 0.072 | 0.0014 | No |
| 37 | Tex15 | 7796 | 0.071 | 0.0032 | No |
| 38 | Cdkal1 | 8063 | 0.060 | -0.0082 | No |
| 39 | Atp4a | 8137 | 0.056 | -0.0099 | No |
| 40 | Sidt1 | 8189 | 0.053 | -0.0106 | No |
| 41 | Gpr19 | 8230 | 0.052 | -0.0108 | No |
| 42 | Bard1 | 8360 | 0.046 | -0.0157 | No |
| 43 | Hsd11b2 | 8406 | 0.044 | -0.0164 | No |
| 44 | Col2a1 | 8981 | 0.020 | -0.0449 | No |
| 45 | Vps50 | 9139 | 0.014 | -0.0523 | No |
| 46 | Tgfb2 | 9361 | 0.004 | -0.0634 | No |
| 47 | Hc | 9766 | -0.009 | -0.0836 | No |
| 48 | Asb7 | 9837 | -0.012 | -0.0868 | No |
| 49 | Cpeb3 | 9876 | -0.013 | -0.0882 | No |
| 50 | Thnsl2 | 9888 | -0.014 | -0.0883 | No |
| 51 | Itgb1bp2 | 10069 | -0.022 | -0.0967 | No |
| 52 | Gamt | 10391 | -0.035 | -0.1117 | No |
| 53 | Bmpr1b | 10556 | -0.042 | -0.1185 | No |
| 54 | Arpp21 | 10653 | -0.046 | -0.1218 | No |
| 55 | Abcg4 | 11197 | -0.067 | -0.1470 | No |
| 56 | Cpb1 | 11657 | -0.087 | -0.1672 | No |
| 57 | Entpd7 | 11777 | -0.092 | -0.1700 | No |
| 58 | Macroh2a2 | 11875 | -0.096 | -0.1715 | No |
| 59 | Ryr1 | 11920 | -0.098 | -0.1702 | No |
| 60 | Cyp39a1 | 12019 | -0.102 | -0.1715 | No |
| 61 | Tnni3 | 13294 | -0.157 | -0.2306 | No |
| 62 | Fggy | 13717 | -0.176 | -0.2458 | No |
| 63 | Prodh | 13718 | -0.176 | -0.2395 | No |
| 64 | Mfsd6 | 13891 | -0.183 | -0.2416 | No |
| 65 | Coq8a | 14008 | -0.189 | -0.2408 | No |
| 66 | Slc16a7 | 14329 | -0.204 | -0.2497 | No |
| 67 | Rsad2 | 14676 | -0.221 | -0.2594 | Yes |
| 68 | Gtf3c5 | 14693 | -0.222 | -0.2523 | Yes |
| 69 | Dcc | 14837 | -0.229 | -0.2513 | Yes |
| 70 | Mthfr | 14947 | -0.233 | -0.2485 | Yes |
| 71 | Kmt2d | 14963 | -0.234 | -0.2409 | Yes |
| 72 | Copz2 | 15221 | -0.248 | -0.2451 | Yes |
| 73 | Efhd1 | 15326 | -0.253 | -0.2413 | Yes |
| 74 | Dtnb | 15527 | -0.264 | -0.2420 | Yes |
| 75 | Kcnn1 | 15528 | -0.264 | -0.2325 | Yes |
| 76 | Arhgdig | 15695 | -0.272 | -0.2312 | Yes |
| 77 | Msh5 | 16080 | -0.295 | -0.2402 | Yes |
| 78 | Plag1 | 16153 | -0.299 | -0.2331 | Yes |
| 79 | Gp1ba | 16289 | -0.308 | -0.2290 | Yes |
| 80 | Idua | 16355 | -0.312 | -0.2211 | Yes |
| 81 | Snn | 16372 | -0.313 | -0.2107 | Yes |
| 82 | Celsr2 | 16400 | -0.314 | -0.2008 | Yes |
| 83 | Zbtb16 | 16407 | -0.315 | -0.1898 | Yes |
| 84 | Egf | 16609 | -0.327 | -0.1883 | Yes |
| 85 | Thrb | 16825 | -0.344 | -0.1869 | Yes |
| 86 | Btg2 | 16851 | -0.346 | -0.1758 | Yes |
| 87 | Serpinb2 | 17025 | -0.358 | -0.1718 | Yes |
| 88 | Ryr2 | 17116 | -0.364 | -0.1633 | Yes |
| 89 | Cpa2 | 17453 | -0.389 | -0.1665 | Yes |
| 90 | Klk8 | 17792 | -0.418 | -0.1687 | Yes |
| 91 | Nr4a2 | 17968 | -0.436 | -0.1620 | Yes |
| 92 | Ypel1 | 18147 | -0.452 | -0.1548 | Yes |
| 93 | Selenop | 18175 | -0.455 | -0.1399 | Yes |
| 94 | Brdt | 18389 | -0.481 | -0.1335 | Yes |
| 95 | Ybx2 | 18676 | -0.523 | -0.1293 | Yes |
| 96 | Prkn | 18685 | -0.525 | -0.1109 | Yes |
| 97 | Mefv | 18889 | -0.557 | -0.1013 | Yes |
| 98 | Mast3 | 18964 | -0.571 | -0.0845 | Yes |
| 99 | Sphk2 | 18969 | -0.572 | -0.0643 | Yes |
| 100 | Kcnd1 | 19272 | -0.650 | -0.0563 | Yes |
| 101 | Ccdc106 | 19383 | -0.686 | -0.0373 | Yes |
| 102 | Synpo | 19493 | -0.731 | -0.0167 | Yes |
| 103 | Slc25a23 | 19643 | -0.858 | 0.0065 | Yes |