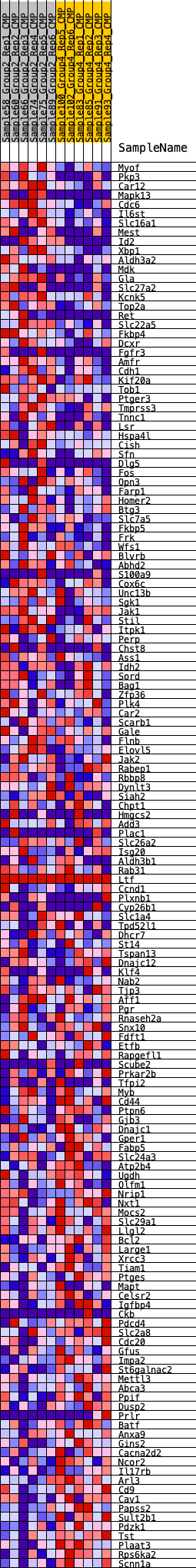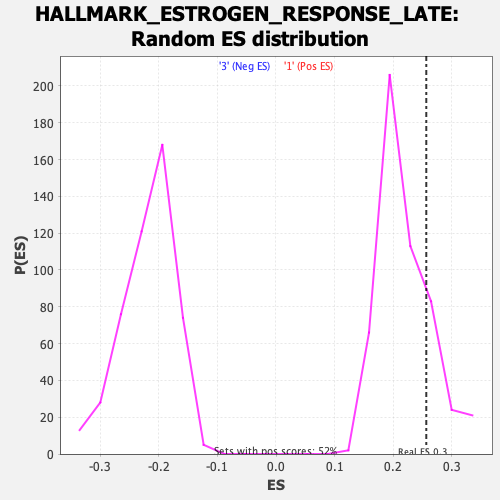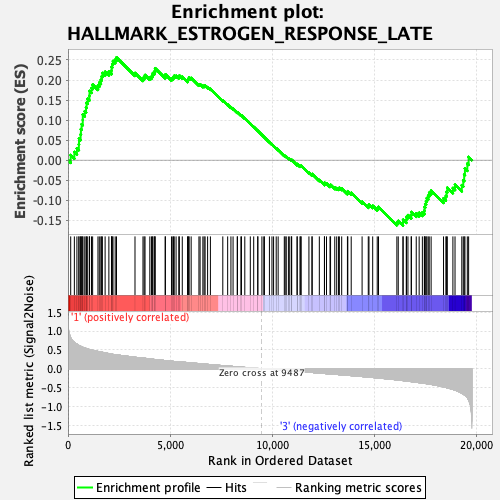
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group4.CMP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_ESTROGEN_RESPONSE_LATE |
| Enrichment Score (ES) | 0.25642687 |
| Normalized Enrichment Score (NES) | 1.1780487 |
| Nominal p-value | 0.18252428 |
| FDR q-value | 0.54109085 |
| FWER p-Value | 0.849 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Myof | 132 | 0.818 | 0.0128 | Yes |
| 2 | Pkp3 | 311 | 0.701 | 0.0204 | Yes |
| 3 | Car12 | 436 | 0.651 | 0.0296 | Yes |
| 4 | Mapk13 | 531 | 0.618 | 0.0395 | Yes |
| 5 | Cdc6 | 539 | 0.616 | 0.0538 | Yes |
| 6 | Il6st | 617 | 0.595 | 0.0640 | Yes |
| 7 | Slc16a1 | 632 | 0.592 | 0.0774 | Yes |
| 8 | Mest | 667 | 0.582 | 0.0896 | Yes |
| 9 | Id2 | 722 | 0.568 | 0.1003 | Yes |
| 10 | Xbp1 | 725 | 0.567 | 0.1138 | Yes |
| 11 | Aldh3a2 | 821 | 0.549 | 0.1220 | Yes |
| 12 | Mdk | 884 | 0.538 | 0.1317 | Yes |
| 13 | Gla | 909 | 0.536 | 0.1432 | Yes |
| 14 | Slc27a2 | 964 | 0.527 | 0.1530 | Yes |
| 15 | Kcnk5 | 1049 | 0.508 | 0.1608 | Yes |
| 16 | Top2a | 1055 | 0.507 | 0.1726 | Yes |
| 17 | Ret | 1150 | 0.496 | 0.1797 | Yes |
| 18 | Slc22a5 | 1206 | 0.489 | 0.1885 | Yes |
| 19 | Fkbp4 | 1467 | 0.458 | 0.1862 | Yes |
| 20 | Dcxr | 1532 | 0.452 | 0.1937 | Yes |
| 21 | Fgfr3 | 1609 | 0.444 | 0.2004 | Yes |
| 22 | Amfr | 1647 | 0.439 | 0.2089 | Yes |
| 23 | Cdh1 | 1683 | 0.434 | 0.2175 | Yes |
| 24 | Kif20a | 1817 | 0.420 | 0.2207 | Yes |
| 25 | Tob1 | 2001 | 0.401 | 0.2209 | Yes |
| 26 | Ptger3 | 2129 | 0.388 | 0.2237 | Yes |
| 27 | Tmprss3 | 2135 | 0.387 | 0.2327 | Yes |
| 28 | Tnnc1 | 2169 | 0.383 | 0.2401 | Yes |
| 29 | Lsr | 2203 | 0.380 | 0.2475 | Yes |
| 30 | Hspa4l | 2310 | 0.372 | 0.2509 | Yes |
| 31 | Cish | 2374 | 0.366 | 0.2564 | Yes |
| 32 | Sfn | 3280 | 0.304 | 0.2175 | No |
| 33 | Dlg5 | 3671 | 0.280 | 0.2043 | No |
| 34 | Fos | 3727 | 0.278 | 0.2082 | No |
| 35 | Opn3 | 3771 | 0.276 | 0.2125 | No |
| 36 | Farp1 | 4006 | 0.260 | 0.2068 | No |
| 37 | Homer2 | 4086 | 0.255 | 0.2088 | No |
| 38 | Btg3 | 4129 | 0.252 | 0.2127 | No |
| 39 | Slc7a5 | 4156 | 0.250 | 0.2173 | No |
| 40 | Fkbp5 | 4227 | 0.245 | 0.2196 | No |
| 41 | Frk | 4257 | 0.243 | 0.2239 | No |
| 42 | Wfs1 | 4265 | 0.242 | 0.2293 | No |
| 43 | Blvrb | 4757 | 0.215 | 0.2094 | No |
| 44 | Abhd2 | 4769 | 0.214 | 0.2139 | No |
| 45 | S100a9 | 5066 | 0.198 | 0.2036 | No |
| 46 | Cox6c | 5131 | 0.195 | 0.2050 | No |
| 47 | Unc13b | 5168 | 0.193 | 0.2077 | No |
| 48 | Sgk1 | 5200 | 0.191 | 0.2107 | No |
| 49 | Jak1 | 5292 | 0.188 | 0.2105 | No |
| 50 | Stil | 5425 | 0.182 | 0.2081 | No |
| 51 | Itpk1 | 5452 | 0.181 | 0.2111 | No |
| 52 | Perp | 5595 | 0.173 | 0.2080 | No |
| 53 | Chst8 | 5853 | 0.163 | 0.1988 | No |
| 54 | Ass1 | 5874 | 0.162 | 0.2016 | No |
| 55 | Idh2 | 5907 | 0.160 | 0.2038 | No |
| 56 | Sord | 5929 | 0.159 | 0.2065 | No |
| 57 | Bag1 | 6030 | 0.153 | 0.2051 | No |
| 58 | Zfp36 | 6411 | 0.136 | 0.1890 | No |
| 59 | Plk4 | 6467 | 0.133 | 0.1893 | No |
| 60 | Car2 | 6604 | 0.127 | 0.1854 | No |
| 61 | Scarb1 | 6663 | 0.123 | 0.1854 | No |
| 62 | Gale | 6712 | 0.121 | 0.1858 | No |
| 63 | Flnb | 6835 | 0.115 | 0.1823 | No |
| 64 | Elovl5 | 6972 | 0.108 | 0.1780 | No |
| 65 | Jak2 | 7579 | 0.081 | 0.1490 | No |
| 66 | Rabep1 | 7817 | 0.071 | 0.1386 | No |
| 67 | Rbbp8 | 7977 | 0.064 | 0.1320 | No |
| 68 | Dynlt3 | 8082 | 0.059 | 0.1281 | No |
| 69 | Siah2 | 8285 | 0.050 | 0.1190 | No |
| 70 | Chpt1 | 8298 | 0.049 | 0.1196 | No |
| 71 | Hmgcs2 | 8466 | 0.042 | 0.1121 | No |
| 72 | Add3 | 8493 | 0.040 | 0.1117 | No |
| 73 | Plac1 | 8656 | 0.034 | 0.1042 | No |
| 74 | Slc26a2 | 8926 | 0.022 | 0.0911 | No |
| 75 | Isg20 | 9092 | 0.016 | 0.0830 | No |
| 76 | Aldh3b1 | 9291 | 0.008 | 0.0731 | No |
| 77 | Rab31 | 9294 | 0.007 | 0.0732 | No |
| 78 | Ltf | 9494 | 0.000 | 0.0630 | No |
| 79 | Ccnd1 | 9586 | -0.001 | 0.0584 | No |
| 80 | Plxnb1 | 9618 | -0.003 | 0.0569 | No |
| 81 | Cyp26b1 | 9862 | -0.013 | 0.0448 | No |
| 82 | Slc1a4 | 9986 | -0.018 | 0.0390 | No |
| 83 | Tpd52l1 | 10065 | -0.022 | 0.0355 | No |
| 84 | Dhcr7 | 10194 | -0.027 | 0.0296 | No |
| 85 | St14 | 10289 | -0.031 | 0.0256 | No |
| 86 | Tspan13 | 10590 | -0.044 | 0.0113 | No |
| 87 | Dnajc12 | 10631 | -0.045 | 0.0104 | No |
| 88 | Klf4 | 10686 | -0.047 | 0.0087 | No |
| 89 | Nab2 | 10791 | -0.052 | 0.0047 | No |
| 90 | Tjp3 | 10856 | -0.054 | 0.0027 | No |
| 91 | Aff1 | 10940 | -0.058 | -0.0001 | No |
| 92 | Pgr | 10952 | -0.059 | 0.0007 | No |
| 93 | Rnaseh2a | 11217 | -0.068 | -0.0111 | No |
| 94 | Snx10 | 11221 | -0.068 | -0.0097 | No |
| 95 | Fdft1 | 11358 | -0.074 | -0.0148 | No |
| 96 | Etfb | 11392 | -0.075 | -0.0147 | No |
| 97 | Rapgefl1 | 11415 | -0.076 | -0.0140 | No |
| 98 | Scube2 | 11796 | -0.092 | -0.0312 | No |
| 99 | Prkar2b | 11943 | -0.099 | -0.0363 | No |
| 100 | Tfpi2 | 11956 | -0.099 | -0.0345 | No |
| 101 | Myb | 12305 | -0.115 | -0.0495 | No |
| 102 | Cd44 | 12556 | -0.126 | -0.0593 | No |
| 103 | Ptpn6 | 12557 | -0.126 | -0.0563 | No |
| 104 | Gjb3 | 12657 | -0.130 | -0.0582 | No |
| 105 | Dnajc1 | 12829 | -0.138 | -0.0637 | No |
| 106 | Gper1 | 12855 | -0.140 | -0.0616 | No |
| 107 | Fabp5 | 13061 | -0.147 | -0.0685 | No |
| 108 | Slc24a3 | 13156 | -0.151 | -0.0697 | No |
| 109 | Atp2b4 | 13246 | -0.156 | -0.0706 | No |
| 110 | Ugdh | 13286 | -0.157 | -0.0688 | No |
| 111 | Olfm1 | 13400 | -0.162 | -0.0707 | No |
| 112 | Nrip1 | 13681 | -0.174 | -0.0808 | No |
| 113 | Nxt1 | 13700 | -0.175 | -0.0776 | No |
| 114 | Mocs2 | 13866 | -0.182 | -0.0816 | No |
| 115 | Slc29a1 | 14399 | -0.207 | -0.1038 | No |
| 116 | Llgl2 | 14701 | -0.222 | -0.1139 | No |
| 117 | Bcl2 | 14736 | -0.223 | -0.1103 | No |
| 118 | Large1 | 14918 | -0.232 | -0.1140 | No |
| 119 | Xrcc3 | 15140 | -0.244 | -0.1194 | No |
| 120 | Tiam1 | 15203 | -0.247 | -0.1167 | No |
| 121 | Ptges | 16102 | -0.296 | -0.1554 | No |
| 122 | Mapt | 16169 | -0.300 | -0.1517 | No |
| 123 | Celsr2 | 16400 | -0.314 | -0.1559 | No |
| 124 | Igfbp4 | 16408 | -0.315 | -0.1487 | No |
| 125 | Ckb | 16564 | -0.325 | -0.1489 | No |
| 126 | Pdcd4 | 16571 | -0.325 | -0.1415 | No |
| 127 | Slc2a8 | 16644 | -0.330 | -0.1373 | No |
| 128 | Cdc20 | 16804 | -0.342 | -0.1372 | No |
| 129 | Gfus | 16813 | -0.343 | -0.1295 | No |
| 130 | Impa2 | 17050 | -0.360 | -0.1329 | No |
| 131 | St6galnac2 | 17193 | -0.369 | -0.1314 | No |
| 132 | Mettl3 | 17350 | -0.380 | -0.1302 | No |
| 133 | Abca3 | 17446 | -0.388 | -0.1258 | No |
| 134 | Ppif | 17457 | -0.389 | -0.1171 | No |
| 135 | Dusp2 | 17489 | -0.391 | -0.1094 | No |
| 136 | Prlr | 17531 | -0.394 | -0.1021 | No |
| 137 | Batf | 17571 | -0.397 | -0.0946 | No |
| 138 | Anxa9 | 17636 | -0.402 | -0.0883 | No |
| 139 | Gins2 | 17686 | -0.408 | -0.0811 | No |
| 140 | Cacna2d2 | 17779 | -0.417 | -0.0759 | No |
| 141 | Ncor2 | 18386 | -0.480 | -0.0953 | No |
| 142 | Il17rb | 18502 | -0.497 | -0.0893 | No |
| 143 | Arl3 | 18536 | -0.503 | -0.0790 | No |
| 144 | Cd9 | 18571 | -0.507 | -0.0687 | No |
| 145 | Cav1 | 18845 | -0.549 | -0.0695 | No |
| 146 | Papss2 | 18949 | -0.568 | -0.0612 | No |
| 147 | Sult2b1 | 19281 | -0.653 | -0.0626 | No |
| 148 | Pdzk1 | 19352 | -0.676 | -0.0500 | No |
| 149 | Tst | 19398 | -0.690 | -0.0359 | No |
| 150 | Plaat3 | 19434 | -0.704 | -0.0209 | No |
| 151 | Rps6ka2 | 19551 | -0.763 | -0.0086 | No |
| 152 | Scnn1a | 19615 | -0.830 | 0.0080 | No |