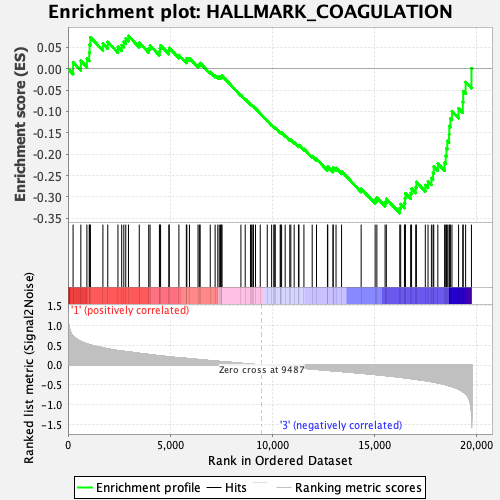
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group4.CMP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.3378536 |
| Normalized Enrichment Score (NES) | -1.2813767 |
| Nominal p-value | 0.099029124 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.763 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pros1 | 249 | 0.727 | 0.0152 | No |
| 2 | Hpn | 629 | 0.593 | 0.0186 | No |
| 3 | Adam9 | 925 | 0.533 | 0.0240 | No |
| 4 | Ang | 1040 | 0.510 | 0.0377 | No |
| 5 | Prep | 1057 | 0.507 | 0.0563 | No |
| 6 | C1qa | 1098 | 0.502 | 0.0735 | No |
| 7 | Olr1 | 1709 | 0.433 | 0.0590 | No |
| 8 | Vwf | 1947 | 0.406 | 0.0625 | No |
| 9 | Mmp9 | 2446 | 0.361 | 0.0510 | No |
| 10 | Fn1 | 2629 | 0.349 | 0.0551 | No |
| 11 | Cfb | 2733 | 0.342 | 0.0630 | No |
| 12 | F10 | 2828 | 0.336 | 0.0710 | No |
| 13 | P2ry1 | 2961 | 0.326 | 0.0768 | No |
| 14 | Comp | 3489 | 0.291 | 0.0612 | No |
| 15 | C9 | 3952 | 0.264 | 0.0478 | No |
| 16 | Gda | 4017 | 0.259 | 0.0544 | No |
| 17 | Capn2 | 4476 | 0.230 | 0.0399 | No |
| 18 | Lgmn | 4502 | 0.229 | 0.0474 | No |
| 19 | Mmp14 | 4533 | 0.227 | 0.0546 | No |
| 20 | Plau | 4937 | 0.204 | 0.0419 | No |
| 21 | C2 | 4958 | 0.204 | 0.0487 | No |
| 22 | Serpinc1 | 5430 | 0.182 | 0.0317 | No |
| 23 | Cpn1 | 5793 | 0.165 | 0.0196 | No |
| 24 | Clu | 5819 | 0.164 | 0.0246 | No |
| 25 | Sirt2 | 5946 | 0.158 | 0.0243 | No |
| 26 | Maff | 6367 | 0.137 | 0.0082 | No |
| 27 | Fyn | 6430 | 0.135 | 0.0102 | No |
| 28 | Mmp8 | 6471 | 0.133 | 0.0132 | No |
| 29 | F3 | 6965 | 0.108 | -0.0077 | No |
| 30 | Lamp2 | 7203 | 0.098 | -0.0160 | No |
| 31 | Mmp2 | 7335 | 0.091 | -0.0191 | No |
| 32 | Fbn1 | 7407 | 0.089 | -0.0194 | No |
| 33 | Plek | 7468 | 0.086 | -0.0191 | No |
| 34 | C3 | 7529 | 0.083 | -0.0190 | No |
| 35 | Timp3 | 7535 | 0.083 | -0.0161 | No |
| 36 | Hmgcs2 | 8466 | 0.042 | -0.0618 | No |
| 37 | Ctso | 8676 | 0.033 | -0.0711 | No |
| 38 | Wdr1 | 8934 | 0.022 | -0.0834 | No |
| 39 | Klkb1 | 8979 | 0.020 | -0.0848 | No |
| 40 | Rapgef3 | 9051 | 0.017 | -0.0878 | No |
| 41 | Pef1 | 9079 | 0.016 | -0.0885 | No |
| 42 | Mmp15 | 9181 | 0.012 | -0.0932 | No |
| 43 | Rac1 | 9412 | 0.003 | -0.1048 | No |
| 44 | Gp9 | 9756 | -0.009 | -0.1219 | No |
| 45 | Lta4h | 9970 | -0.018 | -0.1320 | No |
| 46 | Dusp6 | 10075 | -0.022 | -0.1365 | No |
| 47 | Lrp1 | 10126 | -0.024 | -0.1381 | No |
| 48 | Csrp1 | 10144 | -0.025 | -0.1380 | No |
| 49 | F8 | 10388 | -0.035 | -0.1490 | No |
| 50 | Itgb3 | 10415 | -0.037 | -0.1489 | No |
| 51 | Crip2 | 10458 | -0.038 | -0.1496 | No |
| 52 | Gsn | 10633 | -0.045 | -0.1567 | No |
| 53 | Trf | 10858 | -0.054 | -0.1660 | No |
| 54 | Klf7 | 10910 | -0.057 | -0.1664 | No |
| 55 | Furin | 11073 | -0.064 | -0.1722 | No |
| 56 | S100a1 | 11284 | -0.071 | -0.1801 | No |
| 57 | Arf4 | 11309 | -0.072 | -0.1786 | No |
| 58 | Ctsb | 11551 | -0.082 | -0.1877 | No |
| 59 | Tfpi2 | 11956 | -0.099 | -0.2044 | No |
| 60 | Thbd | 12167 | -0.109 | -0.2110 | No |
| 61 | S100a13 | 12703 | -0.133 | -0.2331 | No |
| 62 | Mmp11 | 12719 | -0.133 | -0.2287 | No |
| 63 | Sparc | 12976 | -0.145 | -0.2362 | No |
| 64 | Ctsl | 12982 | -0.145 | -0.2309 | No |
| 65 | Gnb2 | 13123 | -0.150 | -0.2323 | No |
| 66 | Msrb2 | 13394 | -0.162 | -0.2398 | No |
| 67 | Thbs1 | 14353 | -0.205 | -0.2807 | No |
| 68 | Pecam1 | 15038 | -0.238 | -0.3063 | No |
| 69 | Plg | 15122 | -0.243 | -0.3013 | No |
| 70 | C8g | 15525 | -0.264 | -0.3116 | No |
| 71 | Apoc1 | 15586 | -0.267 | -0.3045 | No |
| 72 | Rabif | 16244 | -0.305 | -0.3262 | Yes |
| 73 | Gp1ba | 16289 | -0.308 | -0.3167 | Yes |
| 74 | Iscu | 16473 | -0.320 | -0.3137 | Yes |
| 75 | Cfd | 16499 | -0.321 | -0.3027 | Yes |
| 76 | Pdgfb | 16521 | -0.323 | -0.2914 | Yes |
| 77 | Ctse | 16776 | -0.340 | -0.2913 | Yes |
| 78 | Itga2 | 16821 | -0.343 | -0.2804 | Yes |
| 79 | Serpinb2 | 17025 | -0.358 | -0.2771 | Yes |
| 80 | Hnf4a | 17058 | -0.360 | -0.2649 | Yes |
| 81 | Proz | 17494 | -0.392 | -0.2720 | Yes |
| 82 | Gng12 | 17627 | -0.401 | -0.2634 | Yes |
| 83 | Klk8 | 17792 | -0.418 | -0.2557 | Yes |
| 84 | Anxa1 | 17872 | -0.425 | -0.2435 | Yes |
| 85 | Usp11 | 17908 | -0.429 | -0.2289 | Yes |
| 86 | Mst1 | 18107 | -0.448 | -0.2218 | Yes |
| 87 | Capn5 | 18438 | -0.488 | -0.2199 | Yes |
| 88 | Casp9 | 18493 | -0.495 | -0.2037 | Yes |
| 89 | Sh2b2 | 18531 | -0.502 | -0.1864 | Yes |
| 90 | Cd9 | 18571 | -0.507 | -0.1689 | Yes |
| 91 | Ctsk | 18657 | -0.521 | -0.1533 | Yes |
| 92 | Ctsh | 18670 | -0.523 | -0.1339 | Yes |
| 93 | F2rl2 | 18724 | -0.530 | -0.1163 | Yes |
| 94 | Cpq | 18799 | -0.541 | -0.0994 | Yes |
| 95 | C8b | 19123 | -0.605 | -0.0927 | Yes |
| 96 | Masp2 | 19331 | -0.669 | -0.0776 | Yes |
| 97 | Bmp1 | 19349 | -0.675 | -0.0526 | Yes |
| 98 | Dpp4 | 19468 | -0.717 | -0.0311 | Yes |
| 99 | Pf4 | 19753 | -1.215 | 0.0009 | Yes |

