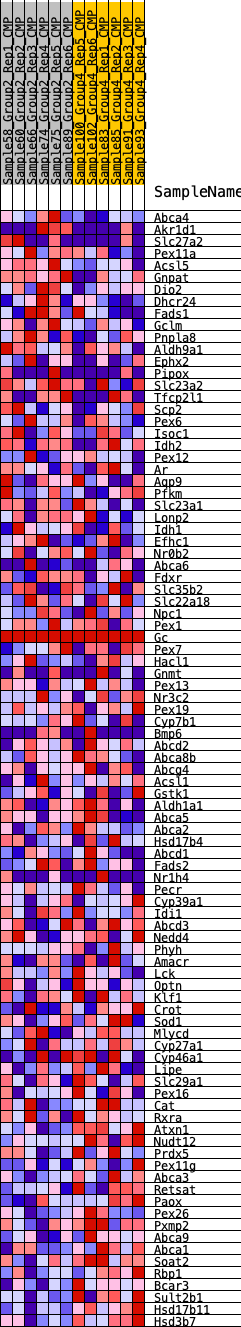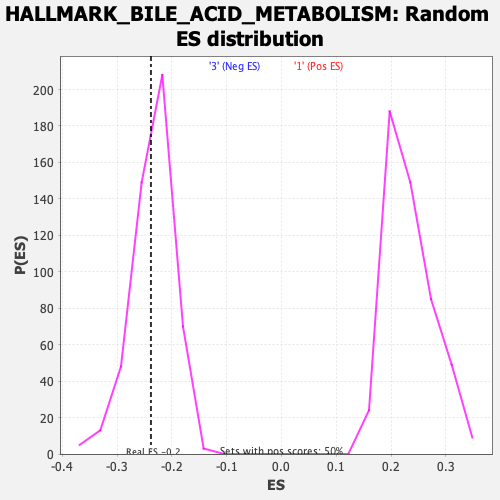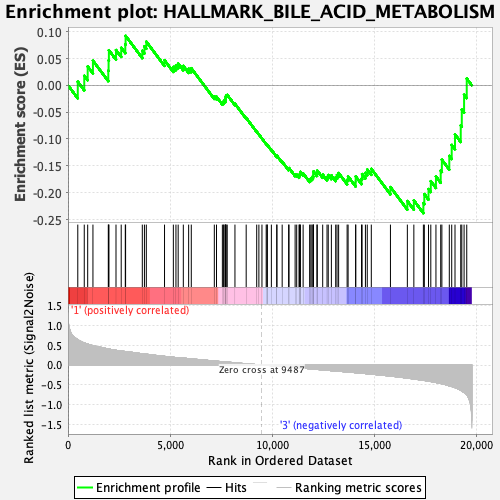
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group4.CMP_Pheno.cls#Group2_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.23812674 |
| Normalized Enrichment Score (NES) | -1.0128633 |
| Nominal p-value | 0.43145162 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.982 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca4 | 479 | 0.636 | 0.0068 | No |
| 2 | Akr1d1 | 796 | 0.554 | 0.0178 | No |
| 3 | Slc27a2 | 964 | 0.527 | 0.0351 | No |
| 4 | Pex11a | 1221 | 0.486 | 0.0459 | No |
| 5 | Acsl5 | 1973 | 0.403 | 0.0275 | No |
| 6 | Gnpat | 1988 | 0.402 | 0.0464 | No |
| 7 | Dio2 | 2002 | 0.401 | 0.0654 | No |
| 8 | Dhcr24 | 2350 | 0.368 | 0.0658 | No |
| 9 | Fads1 | 2605 | 0.350 | 0.0700 | No |
| 10 | Gclm | 2805 | 0.337 | 0.0764 | No |
| 11 | Pnpla8 | 2820 | 0.336 | 0.0921 | No |
| 12 | Aldh9a1 | 3638 | 0.282 | 0.0644 | No |
| 13 | Ephx2 | 3742 | 0.278 | 0.0728 | No |
| 14 | Pipox | 3834 | 0.271 | 0.0814 | No |
| 15 | Slc23a2 | 4726 | 0.217 | 0.0468 | No |
| 16 | Tfcp2l1 | 5158 | 0.193 | 0.0343 | No |
| 17 | Scp2 | 5285 | 0.189 | 0.0371 | No |
| 18 | Pex6 | 5394 | 0.184 | 0.0406 | No |
| 19 | Isoc1 | 5646 | 0.170 | 0.0362 | No |
| 20 | Idh2 | 5907 | 0.160 | 0.0309 | No |
| 21 | Pex12 | 6036 | 0.153 | 0.0318 | No |
| 22 | Ar | 7164 | 0.099 | -0.0206 | No |
| 23 | Aqp9 | 7271 | 0.095 | -0.0213 | No |
| 24 | Pfkm | 7558 | 0.082 | -0.0318 | No |
| 25 | Slc23a1 | 7621 | 0.079 | -0.0311 | No |
| 26 | Lonp2 | 7639 | 0.078 | -0.0282 | No |
| 27 | Idh1 | 7705 | 0.075 | -0.0278 | No |
| 28 | Efhc1 | 7711 | 0.075 | -0.0244 | No |
| 29 | Nr0b2 | 7723 | 0.075 | -0.0213 | No |
| 30 | Abca6 | 7743 | 0.074 | -0.0187 | No |
| 31 | Fdxr | 7795 | 0.071 | -0.0178 | No |
| 32 | Slc35b2 | 8174 | 0.054 | -0.0343 | No |
| 33 | Slc22a18 | 8724 | 0.031 | -0.0607 | No |
| 34 | Npc1 | 9239 | 0.010 | -0.0864 | No |
| 35 | Pex1 | 9346 | 0.005 | -0.0915 | No |
| 36 | Gc | 9496 | 0.000 | -0.0991 | No |
| 37 | Pex7 | 9704 | -0.007 | -0.1093 | No |
| 38 | Hacl1 | 9749 | -0.009 | -0.1111 | No |
| 39 | Gnmt | 9757 | -0.009 | -0.1110 | No |
| 40 | Pex13 | 9957 | -0.017 | -0.1202 | No |
| 41 | Nr3c2 | 10223 | -0.028 | -0.1323 | No |
| 42 | Pex19 | 10227 | -0.028 | -0.1311 | No |
| 43 | Cyp7b1 | 10486 | -0.039 | -0.1423 | No |
| 44 | Bmp6 | 10802 | -0.052 | -0.1558 | No |
| 45 | Abcd2 | 10821 | -0.053 | -0.1541 | No |
| 46 | Abca8b | 11120 | -0.066 | -0.1660 | No |
| 47 | Abcg4 | 11197 | -0.067 | -0.1666 | No |
| 48 | Acsl1 | 11312 | -0.072 | -0.1688 | No |
| 49 | Gstk1 | 11333 | -0.073 | -0.1663 | No |
| 50 | Aldh1a1 | 11362 | -0.074 | -0.1641 | No |
| 51 | Abca5 | 11378 | -0.075 | -0.1612 | No |
| 52 | Abca2 | 11512 | -0.080 | -0.1640 | No |
| 53 | Hsd17b4 | 11831 | -0.093 | -0.1756 | No |
| 54 | Abcd1 | 11890 | -0.096 | -0.1738 | No |
| 55 | Fads2 | 11946 | -0.099 | -0.1718 | No |
| 56 | Nr1h4 | 11994 | -0.101 | -0.1692 | No |
| 57 | Pecr | 12017 | -0.102 | -0.1653 | No |
| 58 | Cyp39a1 | 12019 | -0.102 | -0.1604 | No |
| 59 | Idi1 | 12191 | -0.110 | -0.1637 | No |
| 60 | Abcd3 | 12202 | -0.111 | -0.1588 | No |
| 61 | Nedd4 | 12470 | -0.122 | -0.1664 | No |
| 62 | Phyh | 12682 | -0.132 | -0.1706 | No |
| 63 | Amacr | 12742 | -0.134 | -0.1671 | No |
| 64 | Lck | 12895 | -0.142 | -0.1679 | No |
| 65 | Optn | 13107 | -0.149 | -0.1713 | No |
| 66 | Klf1 | 13171 | -0.152 | -0.1670 | No |
| 67 | Crot | 13250 | -0.156 | -0.1634 | No |
| 68 | Sod1 | 13666 | -0.173 | -0.1760 | No |
| 69 | Mlycd | 13714 | -0.176 | -0.1697 | No |
| 70 | Cyp27a1 | 14085 | -0.192 | -0.1791 | No |
| 71 | Cyp46a1 | 14086 | -0.192 | -0.1697 | No |
| 72 | Lipe | 14370 | -0.206 | -0.1740 | No |
| 73 | Slc29a1 | 14399 | -0.207 | -0.1653 | No |
| 74 | Pex16 | 14562 | -0.214 | -0.1631 | No |
| 75 | Cat | 14652 | -0.220 | -0.1569 | No |
| 76 | Rxra | 14850 | -0.230 | -0.1556 | No |
| 77 | Atxn1 | 15787 | -0.278 | -0.1896 | No |
| 78 | Nudt12 | 16615 | -0.328 | -0.2156 | No |
| 79 | Prdx5 | 16933 | -0.352 | -0.2144 | No |
| 80 | Pex11g | 17400 | -0.384 | -0.2193 | Yes |
| 81 | Abca3 | 17446 | -0.388 | -0.2026 | Yes |
| 82 | Retsat | 17651 | -0.403 | -0.1932 | Yes |
| 83 | Paox | 17763 | -0.415 | -0.1786 | Yes |
| 84 | Pex26 | 18015 | -0.440 | -0.1698 | Yes |
| 85 | Pxmp2 | 18248 | -0.464 | -0.1589 | Yes |
| 86 | Abca9 | 18305 | -0.472 | -0.1386 | Yes |
| 87 | Abca1 | 18669 | -0.522 | -0.1315 | Yes |
| 88 | Soat2 | 18787 | -0.538 | -0.1111 | Yes |
| 89 | Rbp1 | 18948 | -0.567 | -0.0914 | Yes |
| 90 | Bcar3 | 19229 | -0.632 | -0.0747 | Yes |
| 91 | Sult2b1 | 19281 | -0.653 | -0.0454 | Yes |
| 92 | Hsd17b11 | 19393 | -0.688 | -0.0173 | Yes |
| 93 | Hsd3b7 | 19523 | -0.746 | 0.0126 | Yes |