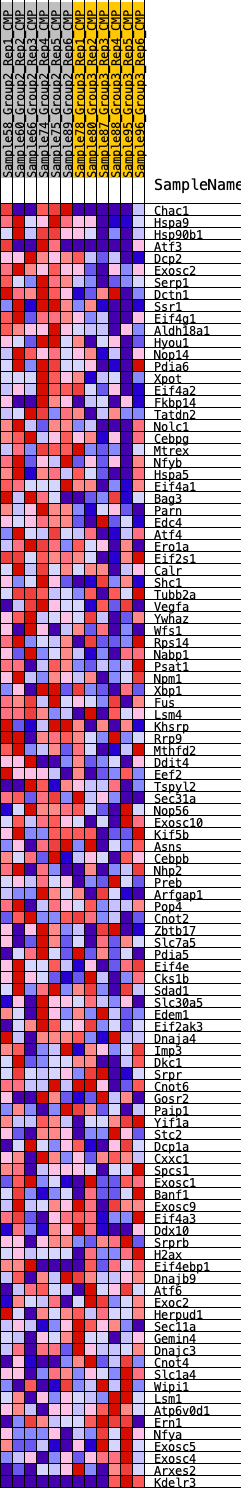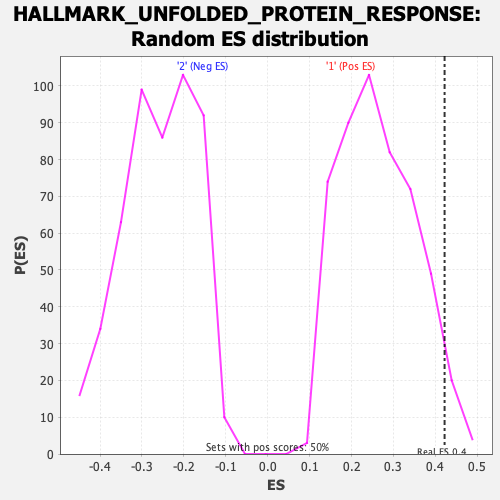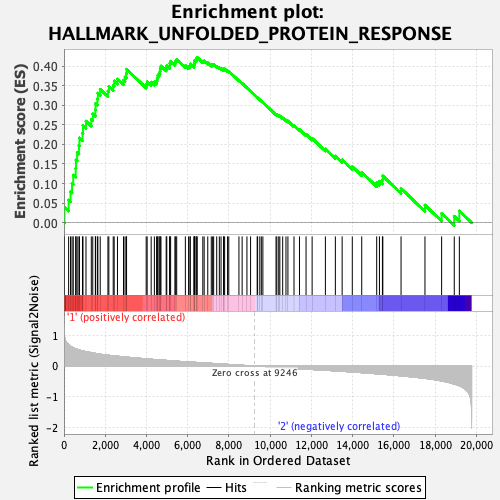
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group3.CMP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.42214227 |
| Normalized Enrichment Score (NES) | 1.5910562 |
| Nominal p-value | 0.04225352 |
| FDR q-value | 0.12582965 |
| FWER p-Value | 0.249 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Chac1 | 22 | 1.003 | 0.0400 | Yes |
| 2 | Hspa9 | 224 | 0.688 | 0.0580 | Yes |
| 3 | Hsp90b1 | 317 | 0.637 | 0.0794 | Yes |
| 4 | Atf3 | 389 | 0.608 | 0.1007 | Yes |
| 5 | Dcp2 | 448 | 0.587 | 0.1218 | Yes |
| 6 | Exosc2 | 567 | 0.555 | 0.1386 | Yes |
| 7 | Serp1 | 581 | 0.551 | 0.1605 | Yes |
| 8 | Dctn1 | 638 | 0.538 | 0.1797 | Yes |
| 9 | Ssr1 | 725 | 0.521 | 0.1967 | Yes |
| 10 | Eif4g1 | 749 | 0.516 | 0.2167 | Yes |
| 11 | Aldh18a1 | 897 | 0.488 | 0.2292 | Yes |
| 12 | Hyou1 | 920 | 0.484 | 0.2479 | Yes |
| 13 | Nop14 | 1066 | 0.464 | 0.2596 | Yes |
| 14 | Pdia6 | 1324 | 0.431 | 0.2642 | Yes |
| 15 | Xpot | 1395 | 0.425 | 0.2780 | Yes |
| 16 | Eif4a2 | 1521 | 0.407 | 0.2883 | Yes |
| 17 | Fkbp14 | 1530 | 0.405 | 0.3045 | Yes |
| 18 | Tatdn2 | 1617 | 0.394 | 0.3163 | Yes |
| 19 | Nolc1 | 1642 | 0.391 | 0.3311 | Yes |
| 20 | Cebpg | 1757 | 0.380 | 0.3409 | Yes |
| 21 | Mtrex | 2131 | 0.349 | 0.3362 | Yes |
| 22 | Nfyb | 2175 | 0.345 | 0.3482 | Yes |
| 23 | Hspa5 | 2387 | 0.330 | 0.3510 | Yes |
| 24 | Eif4a1 | 2433 | 0.326 | 0.3620 | Yes |
| 25 | Bag3 | 2593 | 0.316 | 0.3669 | Yes |
| 26 | Parn | 2886 | 0.296 | 0.3642 | Yes |
| 27 | Edc4 | 2960 | 0.290 | 0.3724 | Yes |
| 28 | Atf4 | 3022 | 0.286 | 0.3810 | Yes |
| 29 | Ero1a | 3034 | 0.285 | 0.3921 | Yes |
| 30 | Eif2s1 | 3986 | 0.229 | 0.3531 | Yes |
| 31 | Calr | 4033 | 0.226 | 0.3601 | Yes |
| 32 | Shc1 | 4228 | 0.218 | 0.3591 | Yes |
| 33 | Tubb2a | 4383 | 0.208 | 0.3598 | Yes |
| 34 | Vegfa | 4493 | 0.202 | 0.3625 | Yes |
| 35 | Ywhaz | 4516 | 0.200 | 0.3696 | Yes |
| 36 | Wfs1 | 4545 | 0.199 | 0.3763 | Yes |
| 37 | Rps14 | 4609 | 0.196 | 0.3812 | Yes |
| 38 | Nabp1 | 4655 | 0.194 | 0.3869 | Yes |
| 39 | Psat1 | 4665 | 0.193 | 0.3943 | Yes |
| 40 | Npm1 | 4703 | 0.192 | 0.4003 | Yes |
| 41 | Xbp1 | 4956 | 0.181 | 0.3949 | Yes |
| 42 | Fus | 4985 | 0.179 | 0.4008 | Yes |
| 43 | Lsm4 | 5124 | 0.172 | 0.4009 | Yes |
| 44 | Khsrp | 5133 | 0.171 | 0.4075 | Yes |
| 45 | Rrp9 | 5172 | 0.170 | 0.4125 | Yes |
| 46 | Mthfd2 | 5379 | 0.160 | 0.4086 | Yes |
| 47 | Ddit4 | 5417 | 0.158 | 0.4132 | Yes |
| 48 | Eef2 | 5466 | 0.155 | 0.4171 | Yes |
| 49 | Tspyl2 | 5886 | 0.139 | 0.4014 | Yes |
| 50 | Sec31a | 6042 | 0.132 | 0.3990 | Yes |
| 51 | Nop56 | 6114 | 0.128 | 0.4006 | Yes |
| 52 | Exosc10 | 6121 | 0.128 | 0.4056 | Yes |
| 53 | Kif5b | 6302 | 0.120 | 0.4013 | Yes |
| 54 | Asns | 6317 | 0.119 | 0.4055 | Yes |
| 55 | Cebpb | 6328 | 0.119 | 0.4099 | Yes |
| 56 | Nhp2 | 6333 | 0.119 | 0.4145 | Yes |
| 57 | Preb | 6409 | 0.115 | 0.4155 | Yes |
| 58 | Arfgap1 | 6412 | 0.115 | 0.4201 | Yes |
| 59 | Pop4 | 6463 | 0.113 | 0.4221 | Yes |
| 60 | Cnot2 | 6733 | 0.102 | 0.4126 | No |
| 61 | Zbtb17 | 6798 | 0.100 | 0.4135 | No |
| 62 | Slc7a5 | 6973 | 0.092 | 0.4084 | No |
| 63 | Pdia5 | 7143 | 0.084 | 0.4033 | No |
| 64 | Eif4e | 7208 | 0.082 | 0.4034 | No |
| 65 | Cks1b | 7259 | 0.079 | 0.4041 | No |
| 66 | Sdad1 | 7407 | 0.073 | 0.3996 | No |
| 67 | Slc30a5 | 7535 | 0.067 | 0.3959 | No |
| 68 | Edem1 | 7617 | 0.064 | 0.3944 | No |
| 69 | Eif2ak3 | 7741 | 0.059 | 0.3906 | No |
| 70 | Dnaja4 | 7743 | 0.059 | 0.3929 | No |
| 71 | Imp3 | 7791 | 0.057 | 0.3929 | No |
| 72 | Dkc1 | 7933 | 0.051 | 0.3878 | No |
| 73 | Srpr | 7992 | 0.049 | 0.3869 | No |
| 74 | Cnot6 | 8485 | 0.028 | 0.3630 | No |
| 75 | Gosr2 | 8640 | 0.021 | 0.3561 | No |
| 76 | Paip1 | 8873 | 0.012 | 0.3448 | No |
| 77 | Yif1a | 9049 | 0.006 | 0.3361 | No |
| 78 | Stc2 | 9369 | -0.003 | 0.3200 | No |
| 79 | Dcp1a | 9392 | -0.003 | 0.3190 | No |
| 80 | Cxxc1 | 9497 | -0.007 | 0.3140 | No |
| 81 | Spcs1 | 9571 | -0.010 | 0.3107 | No |
| 82 | Exosc1 | 9646 | -0.013 | 0.3075 | No |
| 83 | Banf1 | 10284 | -0.038 | 0.2767 | No |
| 84 | Exosc9 | 10324 | -0.040 | 0.2763 | No |
| 85 | Eif4a3 | 10416 | -0.044 | 0.2735 | No |
| 86 | Ddx10 | 10469 | -0.045 | 0.2727 | No |
| 87 | Srprb | 10607 | -0.051 | 0.2678 | No |
| 88 | H2ax | 10766 | -0.058 | 0.2621 | No |
| 89 | Eif4ebp1 | 10861 | -0.062 | 0.2599 | No |
| 90 | Dnajb9 | 11156 | -0.076 | 0.2480 | No |
| 91 | Atf6 | 11422 | -0.086 | 0.2381 | No |
| 92 | Exoc2 | 11740 | -0.098 | 0.2260 | No |
| 93 | Herpud1 | 12035 | -0.112 | 0.2156 | No |
| 94 | Sec11a | 12678 | -0.136 | 0.1885 | No |
| 95 | Gemin4 | 13162 | -0.155 | 0.1703 | No |
| 96 | Dnajc3 | 13491 | -0.170 | 0.1606 | No |
| 97 | Cnot4 | 13983 | -0.190 | 0.1434 | No |
| 98 | Slc1a4 | 14441 | -0.213 | 0.1289 | No |
| 99 | Wipi1 | 15167 | -0.250 | 0.1023 | No |
| 100 | Lsm1 | 15300 | -0.257 | 0.1062 | No |
| 101 | Atp6v0d1 | 15445 | -0.264 | 0.1097 | No |
| 102 | Ern1 | 15464 | -0.265 | 0.1196 | No |
| 103 | Nfya | 16348 | -0.317 | 0.0877 | No |
| 104 | Exosc5 | 17508 | -0.399 | 0.0451 | No |
| 105 | Exosc4 | 18316 | -0.482 | 0.0238 | No |
| 106 | Arxes2 | 18929 | -0.583 | 0.0166 | No |
| 107 | Kdelr3 | 19176 | -0.638 | 0.0303 | No |