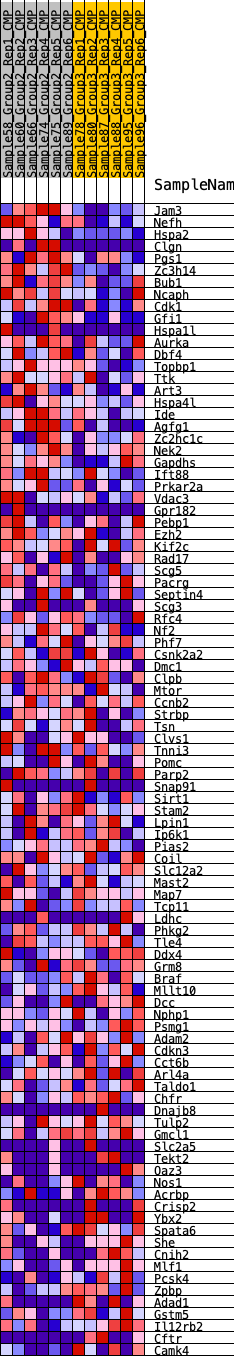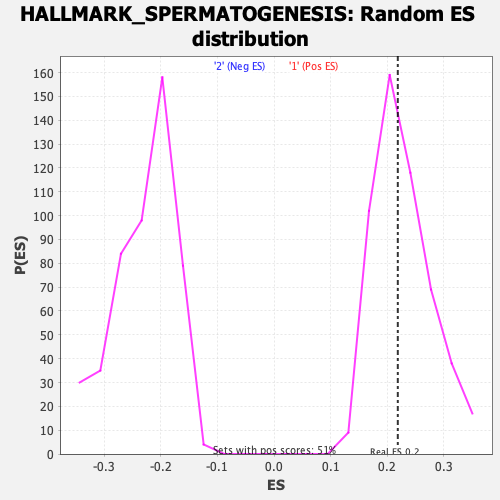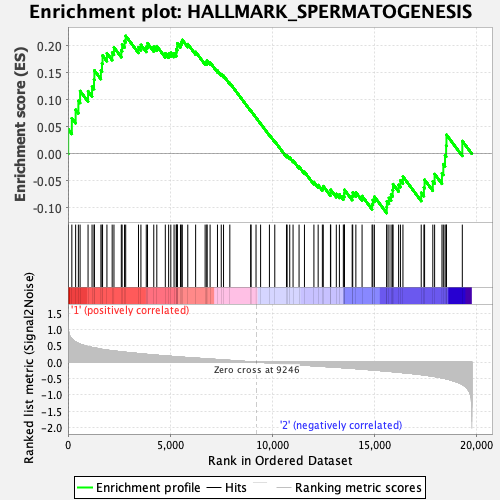
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group3.CMP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.21878648 |
| Normalized Enrichment Score (NES) | 0.9657641 |
| Nominal p-value | 0.50390625 |
| FDR q-value | 0.8686661 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Jam3 | 9 | 1.124 | 0.0456 | Yes |
| 2 | Nefh | 188 | 0.714 | 0.0659 | Yes |
| 3 | Hspa2 | 374 | 0.614 | 0.0816 | Yes |
| 4 | Clgn | 508 | 0.569 | 0.0982 | Yes |
| 5 | Pgs1 | 592 | 0.547 | 0.1165 | Yes |
| 6 | Zc3h14 | 982 | 0.474 | 0.1161 | Yes |
| 7 | Bub1 | 1178 | 0.448 | 0.1246 | Yes |
| 8 | Ncaph | 1274 | 0.438 | 0.1377 | Yes |
| 9 | Cdk1 | 1292 | 0.434 | 0.1546 | Yes |
| 10 | Gfi1 | 1611 | 0.395 | 0.1547 | Yes |
| 11 | Hspa1l | 1665 | 0.389 | 0.1679 | Yes |
| 12 | Aurka | 1692 | 0.386 | 0.1824 | Yes |
| 13 | Dbf4 | 1906 | 0.366 | 0.1866 | Yes |
| 14 | Topbp1 | 2163 | 0.346 | 0.1878 | Yes |
| 15 | Ttk | 2250 | 0.340 | 0.1974 | Yes |
| 16 | Art3 | 2610 | 0.315 | 0.1921 | Yes |
| 17 | Hspa4l | 2649 | 0.312 | 0.2029 | Yes |
| 18 | Ide | 2768 | 0.303 | 0.2094 | Yes |
| 19 | Agfg1 | 2825 | 0.299 | 0.2188 | Yes |
| 20 | Zc2hc1c | 3451 | 0.261 | 0.1977 | No |
| 21 | Nek2 | 3570 | 0.254 | 0.2021 | No |
| 22 | Gapdhs | 3832 | 0.238 | 0.1986 | No |
| 23 | Ift88 | 3893 | 0.234 | 0.2051 | No |
| 24 | Prkar2a | 4199 | 0.219 | 0.1986 | No |
| 25 | Vdac3 | 4348 | 0.209 | 0.1997 | No |
| 26 | Gpr182 | 4764 | 0.189 | 0.1864 | No |
| 27 | Pebp1 | 4927 | 0.182 | 0.1856 | No |
| 28 | Ezh2 | 5034 | 0.177 | 0.1875 | No |
| 29 | Kif2c | 5193 | 0.169 | 0.1864 | No |
| 30 | Rad17 | 5301 | 0.163 | 0.1876 | No |
| 31 | Scg5 | 5305 | 0.163 | 0.1942 | No |
| 32 | Pacrg | 5351 | 0.161 | 0.1985 | No |
| 33 | Septin4 | 5353 | 0.161 | 0.2050 | No |
| 34 | Scg3 | 5507 | 0.153 | 0.2035 | No |
| 35 | Rfc4 | 5545 | 0.151 | 0.2078 | No |
| 36 | Nf2 | 5596 | 0.149 | 0.2114 | No |
| 37 | Phf7 | 5864 | 0.139 | 0.2035 | No |
| 38 | Csnk2a2 | 6249 | 0.122 | 0.1890 | No |
| 39 | Dmc1 | 6714 | 0.103 | 0.1696 | No |
| 40 | Clpb | 6800 | 0.100 | 0.1694 | No |
| 41 | Mtor | 6806 | 0.100 | 0.1732 | No |
| 42 | Ccnb2 | 6957 | 0.093 | 0.1694 | No |
| 43 | Strbp | 7318 | 0.077 | 0.1543 | No |
| 44 | Tsn | 7504 | 0.068 | 0.1477 | No |
| 45 | Clvs1 | 7615 | 0.064 | 0.1447 | No |
| 46 | Tnni3 | 7923 | 0.052 | 0.1312 | No |
| 47 | Pomc | 8945 | 0.010 | 0.0797 | No |
| 48 | Parp2 | 8962 | 0.009 | 0.0793 | No |
| 49 | Snap91 | 9206 | 0.001 | 0.0670 | No |
| 50 | Sirt1 | 9430 | -0.004 | 0.0558 | No |
| 51 | Stam2 | 9862 | -0.021 | 0.0348 | No |
| 52 | Lpin1 | 10124 | -0.032 | 0.0229 | No |
| 53 | Ip6k1 | 10697 | -0.055 | -0.0040 | No |
| 54 | Pias2 | 10728 | -0.056 | -0.0032 | No |
| 55 | Coil | 10856 | -0.062 | -0.0071 | No |
| 56 | Slc12a2 | 11019 | -0.069 | -0.0125 | No |
| 57 | Mast2 | 11313 | -0.082 | -0.0241 | No |
| 58 | Map7 | 11577 | -0.093 | -0.0336 | No |
| 59 | Tcp11 | 12040 | -0.112 | -0.0525 | No |
| 60 | Ldhc | 12248 | -0.120 | -0.0581 | No |
| 61 | Phkg2 | 12454 | -0.128 | -0.0633 | No |
| 62 | Tle4 | 12498 | -0.130 | -0.0601 | No |
| 63 | Ddx4 | 12850 | -0.142 | -0.0722 | No |
| 64 | Grm8 | 12863 | -0.142 | -0.0669 | No |
| 65 | Braf | 13137 | -0.153 | -0.0745 | No |
| 66 | Mllt10 | 13288 | -0.161 | -0.0756 | No |
| 67 | Dcc | 13481 | -0.169 | -0.0784 | No |
| 68 | Nphp1 | 13527 | -0.172 | -0.0736 | No |
| 69 | Psmg1 | 13535 | -0.172 | -0.0669 | No |
| 70 | Adam2 | 13914 | -0.187 | -0.0785 | No |
| 71 | Cdkn3 | 13940 | -0.188 | -0.0720 | No |
| 72 | Cct6b | 14094 | -0.197 | -0.0717 | No |
| 73 | Arl4a | 14398 | -0.210 | -0.0785 | No |
| 74 | Taldo1 | 14888 | -0.236 | -0.0936 | No |
| 75 | Chfr | 14925 | -0.237 | -0.0857 | No |
| 76 | Dnajb8 | 15001 | -0.242 | -0.0796 | No |
| 77 | Tulp2 | 15605 | -0.272 | -0.0991 | No |
| 78 | Gmcl1 | 15613 | -0.273 | -0.0883 | No |
| 79 | Slc2a5 | 15713 | -0.278 | -0.0819 | No |
| 80 | Tekt2 | 15819 | -0.285 | -0.0756 | No |
| 81 | Oaz3 | 15896 | -0.290 | -0.0675 | No |
| 82 | Nos1 | 15913 | -0.291 | -0.0564 | No |
| 83 | Acrbp | 16187 | -0.307 | -0.0577 | No |
| 84 | Crisp2 | 16276 | -0.312 | -0.0494 | No |
| 85 | Ybx2 | 16399 | -0.320 | -0.0425 | No |
| 86 | Spata6 | 17294 | -0.380 | -0.0723 | No |
| 87 | She | 17426 | -0.391 | -0.0629 | No |
| 88 | Cnih2 | 17459 | -0.394 | -0.0484 | No |
| 89 | Mlf1 | 17862 | -0.429 | -0.0512 | No |
| 90 | Pcsk4 | 17950 | -0.438 | -0.0376 | No |
| 91 | Zpbp | 18310 | -0.481 | -0.0362 | No |
| 92 | Adad1 | 18376 | -0.491 | -0.0193 | No |
| 93 | Gstm5 | 18460 | -0.500 | -0.0031 | No |
| 94 | Il12rb2 | 18514 | -0.508 | 0.0151 | No |
| 95 | Cftr | 18530 | -0.510 | 0.0352 | No |
| 96 | Camk4 | 19306 | -0.678 | 0.0236 | No |