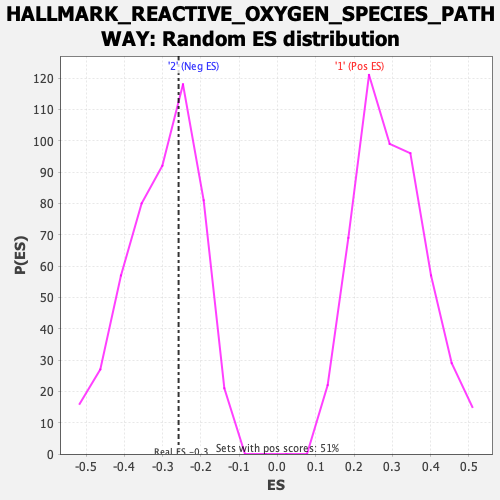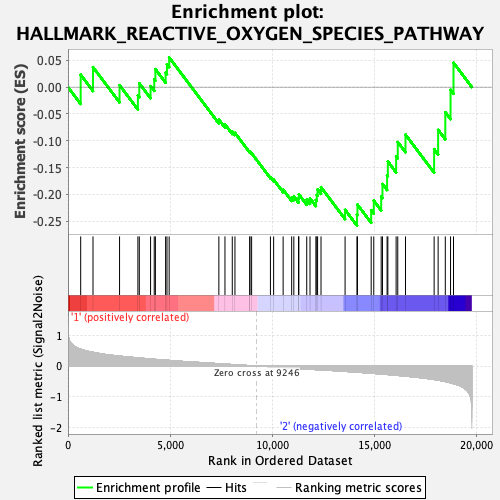
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group3.CMP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY |
| Enrichment Score (ES) | -0.25827497 |
| Normalized Enrichment Score (NES) | -0.8597364 |
| Nominal p-value | 0.6219512 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gpx4 | 621 | 0.542 | 0.0231 | No |
| 2 | Txnrd1 | 1224 | 0.444 | 0.0373 | No |
| 3 | Srxn1 | 2524 | 0.319 | 0.0036 | No |
| 4 | Pfkp | 3422 | 0.262 | -0.0155 | No |
| 5 | Oxsr1 | 3491 | 0.259 | 0.0071 | No |
| 6 | Gclm | 4043 | 0.226 | 0.0019 | No |
| 7 | Sbno2 | 4222 | 0.218 | 0.0148 | No |
| 8 | Mgst1 | 4279 | 0.214 | 0.0336 | No |
| 9 | Hmox2 | 4779 | 0.189 | 0.0273 | No |
| 10 | Glrx2 | 4846 | 0.185 | 0.0426 | No |
| 11 | Prdx1 | 4953 | 0.181 | 0.0554 | No |
| 12 | Gsr | 7386 | 0.073 | -0.0605 | No |
| 13 | Prnp | 7685 | 0.061 | -0.0694 | No |
| 14 | Nqo1 | 8043 | 0.047 | -0.0828 | No |
| 15 | Scaf4 | 8174 | 0.041 | -0.0853 | No |
| 16 | Junb | 8899 | 0.011 | -0.1208 | No |
| 17 | Stk25 | 8905 | 0.011 | -0.1200 | No |
| 18 | Msra | 8990 | 0.008 | -0.1234 | No |
| 19 | G6pdx | 9911 | -0.023 | -0.1677 | No |
| 20 | Abcc1 | 10068 | -0.030 | -0.1725 | No |
| 21 | Ptpa | 10533 | -0.048 | -0.1913 | No |
| 22 | Ndufa6 | 10951 | -0.066 | -0.2058 | No |
| 23 | Mbp | 11057 | -0.071 | -0.2040 | No |
| 24 | Sod2 | 11286 | -0.080 | -0.2074 | No |
| 25 | Glrx | 11303 | -0.081 | -0.2001 | No |
| 26 | Txn1 | 11689 | -0.097 | -0.2098 | No |
| 27 | Prdx6 | 11849 | -0.103 | -0.2075 | No |
| 28 | Prdx4 | 12137 | -0.115 | -0.2105 | No |
| 29 | Ndufs2 | 12182 | -0.117 | -0.2009 | No |
| 30 | Ercc2 | 12220 | -0.119 | -0.1908 | No |
| 31 | Lsp1 | 12388 | -0.126 | -0.1866 | No |
| 32 | Ftl1 | 13566 | -0.173 | -0.2288 | No |
| 33 | Atox1 | 14148 | -0.200 | -0.2381 | Yes |
| 34 | Sod1 | 14172 | -0.201 | -0.2190 | Yes |
| 35 | Gclc | 14844 | -0.234 | -0.2295 | Yes |
| 36 | Prdx2 | 14971 | -0.240 | -0.2116 | Yes |
| 37 | Cat | 15334 | -0.259 | -0.2039 | Yes |
| 38 | Fes | 15397 | -0.262 | -0.1806 | Yes |
| 39 | Cdkn2d | 15623 | -0.273 | -0.1645 | Yes |
| 40 | Mpo | 15660 | -0.275 | -0.1386 | Yes |
| 41 | Pdlim1 | 16065 | -0.299 | -0.1290 | Yes |
| 42 | Egln2 | 16145 | -0.304 | -0.1024 | Yes |
| 43 | Gpx3 | 16524 | -0.328 | -0.0885 | Yes |
| 44 | Lamtor5 | 17926 | -0.436 | -0.1157 | Yes |
| 45 | Ipcef1 | 18122 | -0.456 | -0.0796 | Yes |
| 46 | Selenos | 18473 | -0.503 | -0.0468 | Yes |
| 47 | Txnrd2 | 18730 | -0.544 | -0.0050 | Yes |
| 48 | Hhex | 18875 | -0.573 | 0.0454 | Yes |