Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group3.CMP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
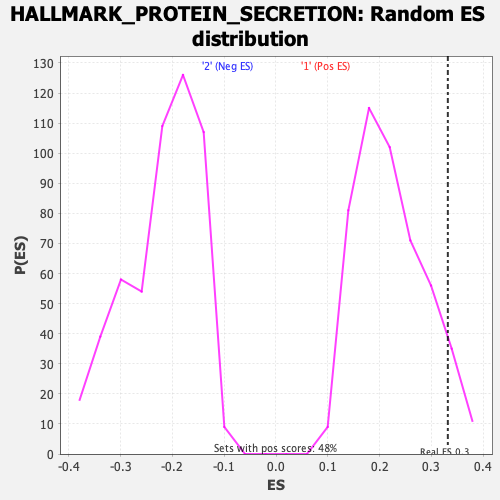
| Enrichment Score (ES) | 0.33171088 |
| Normalized Enrichment Score (NES) | 1.4943849 |
| Nominal p-value | 0.052083332 |
| FDR q-value | 0.16346703 |
| FWER p-Value | 0.409 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 80 | 0.804 | 0.0342 | Yes |
| 2 | Ykt6 | 251 | 0.670 | 0.0575 | Yes |
| 3 | Anp32e | 261 | 0.664 | 0.0886 | Yes |
| 4 | Dst | 290 | 0.651 | 0.1182 | Yes |
| 5 | Cltc | 307 | 0.641 | 0.1479 | Yes |
| 6 | Rps6ka3 | 443 | 0.589 | 0.1691 | Yes |
| 7 | Atp1a1 | 538 | 0.562 | 0.1911 | Yes |
| 8 | Krt18 | 618 | 0.543 | 0.2130 | Yes |
| 9 | Cln5 | 845 | 0.497 | 0.2252 | Yes |
| 10 | Vamp4 | 860 | 0.493 | 0.2479 | Yes |
| 11 | Adam10 | 1135 | 0.453 | 0.2556 | Yes |
| 12 | Arf1 | 1206 | 0.446 | 0.2733 | Yes |
| 13 | Lman1 | 2023 | 0.356 | 0.2488 | Yes |
| 14 | Copb2 | 2097 | 0.350 | 0.2618 | Yes |
| 15 | Gla | 2228 | 0.341 | 0.2714 | Yes |
| 16 | Clcn3 | 2235 | 0.341 | 0.2873 | Yes |
| 17 | Arcn1 | 2401 | 0.329 | 0.2946 | Yes |
| 18 | Vamp3 | 2445 | 0.325 | 0.3079 | Yes |
| 19 | Copb1 | 2553 | 0.318 | 0.3177 | Yes |
| 20 | Arfgef2 | 2791 | 0.301 | 0.3200 | Yes |
| 21 | Tmed10 | 3015 | 0.287 | 0.3223 | Yes |
| 22 | Snx2 | 3232 | 0.274 | 0.3244 | Yes |
| 23 | Uso1 | 3384 | 0.264 | 0.3293 | Yes |
| 24 | Dnm1l | 3575 | 0.254 | 0.3317 | Yes |
| 25 | Igf2r | 3863 | 0.235 | 0.3283 | No |
| 26 | Ap3b1 | 4221 | 0.218 | 0.3206 | No |
| 27 | Rab5a | 4248 | 0.217 | 0.3296 | No |
| 28 | Ap1g1 | 4669 | 0.193 | 0.3174 | No |
| 29 | Lamp2 | 5109 | 0.173 | 0.3034 | No |
| 30 | Cav2 | 5139 | 0.171 | 0.3100 | No |
| 31 | Mapk1 | 5311 | 0.163 | 0.3091 | No |
| 32 | Ap2b1 | 5632 | 0.147 | 0.2998 | No |
| 33 | Arfip1 | 5643 | 0.146 | 0.3063 | No |
| 34 | Vps4b | 5732 | 0.143 | 0.3086 | No |
| 35 | Stx16 | 5885 | 0.139 | 0.3075 | No |
| 36 | M6pr | 5981 | 0.134 | 0.3091 | No |
| 37 | Sec31a | 6042 | 0.132 | 0.3124 | No |
| 38 | Ap2m1 | 6110 | 0.128 | 0.3151 | No |
| 39 | Stx12 | 6128 | 0.128 | 0.3203 | No |
| 40 | Tmx1 | 6290 | 0.120 | 0.3178 | No |
| 41 | Golga4 | 6316 | 0.119 | 0.3222 | No |
| 42 | Arfgef1 | 6405 | 0.115 | 0.3233 | No |
| 43 | Tpd52 | 6524 | 0.110 | 0.3225 | No |
| 44 | Atp6v1h | 6859 | 0.098 | 0.3102 | No |
| 45 | Zw10 | 6919 | 0.095 | 0.3117 | No |
| 46 | Napg | 6991 | 0.091 | 0.3125 | No |
| 47 | Clta | 7001 | 0.091 | 0.3163 | No |
| 48 | Mon2 | 7121 | 0.085 | 0.3144 | No |
| 49 | Sh3gl2 | 7297 | 0.077 | 0.3092 | No |
| 50 | Cog2 | 7525 | 0.068 | 0.3009 | No |
| 51 | Sec22b | 8148 | 0.042 | 0.2713 | No |
| 52 | Ppt1 | 8371 | 0.032 | 0.2615 | No |
| 53 | Gosr2 | 8640 | 0.021 | 0.2489 | No |
| 54 | Rab9 | 8762 | 0.017 | 0.2436 | No |
| 55 | Gbf1 | 9058 | 0.006 | 0.2289 | No |
| 56 | Rab14 | 9362 | -0.002 | 0.2136 | No |
| 57 | Ctsc | 9601 | -0.011 | 0.2020 | No |
| 58 | Ap2s1 | 9856 | -0.021 | 0.1901 | No |
| 59 | Vps45 | 9906 | -0.023 | 0.1887 | No |
| 60 | Tom1l1 | 10275 | -0.038 | 0.1718 | No |
| 61 | Rab2a | 10334 | -0.040 | 0.1708 | No |
| 62 | Atp7a | 10815 | -0.060 | 0.1492 | No |
| 63 | Rab22a | 11223 | -0.078 | 0.1323 | No |
| 64 | Snap23 | 11238 | -0.078 | 0.1353 | No |
| 65 | Pam | 11245 | -0.078 | 0.1387 | No |
| 66 | Scamp1 | 11376 | -0.084 | 0.1361 | No |
| 67 | Arfgap3 | 11439 | -0.087 | 0.1371 | No |
| 68 | Cope | 11749 | -0.099 | 0.1261 | No |
| 69 | Stx7 | 12030 | -0.111 | 0.1172 | No |
| 70 | Ocrl | 12070 | -0.113 | 0.1205 | No |
| 71 | Scamp3 | 12123 | -0.115 | 0.1234 | No |
| 72 | Tsg101 | 12475 | -0.129 | 0.1117 | No |
| 73 | Sec24d | 12506 | -0.130 | 0.1163 | No |
| 74 | Dop1a | 12618 | -0.134 | 0.1171 | No |
| 75 | Stam | 12755 | -0.139 | 0.1168 | No |
| 76 | Sgms1 | 13769 | -0.181 | 0.0739 | No |
| 77 | Ergic3 | 13771 | -0.181 | 0.0825 | No |
| 78 | Sod1 | 14172 | -0.201 | 0.0718 | No |
| 79 | Ap3s1 | 14482 | -0.215 | 0.0663 | No |
| 80 | Rer1 | 14525 | -0.217 | 0.0745 | No |
| 81 | Ica1 | 14682 | -0.225 | 0.0773 | No |
| 82 | Kif1b | 14773 | -0.230 | 0.0837 | No |
| 83 | Egfr | 14999 | -0.242 | 0.0838 | No |
| 84 | Yipf6 | 15823 | -0.285 | 0.0555 | No |
| 85 | Gnas | 15994 | -0.296 | 0.0610 | No |
| 86 | Napa | 16033 | -0.298 | 0.0732 | No |
| 87 | Bet1 | 16258 | -0.311 | 0.0767 | No |
| 88 | Tspan8 | 16570 | -0.331 | 0.0766 | No |
| 89 | Galc | 17355 | -0.386 | 0.0552 | No |
| 90 | Cd63 | 17584 | -0.405 | 0.0629 | No |
| 91 | Bnip3 | 18237 | -0.471 | 0.0522 | No |
| 92 | Scrn1 | 18707 | -0.539 | 0.0541 | No |