Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group3.CMP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
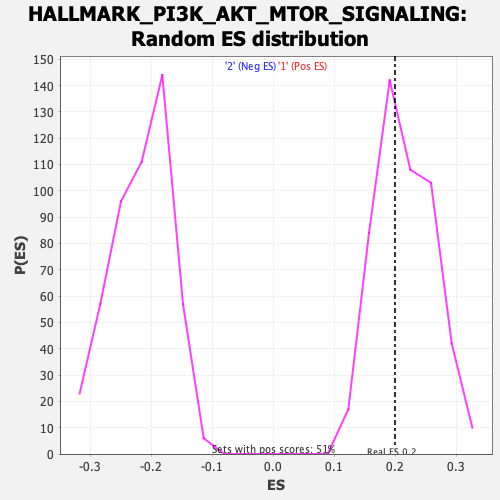
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.19931841 |
| Normalized Enrichment Score (NES) | 0.92445487 |
| Nominal p-value | 0.59881425 |
| FDR q-value | 0.86093825 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cltc | 307 | 0.641 | 0.0147 | Yes |
| 2 | Hsp90b1 | 317 | 0.637 | 0.0443 | Yes |
| 3 | Rps6ka3 | 443 | 0.589 | 0.0657 | Yes |
| 4 | Myd88 | 960 | 0.478 | 0.0620 | Yes |
| 5 | Arf1 | 1206 | 0.446 | 0.0706 | Yes |
| 6 | Cdk1 | 1292 | 0.434 | 0.0868 | Yes |
| 7 | Akt1 | 1543 | 0.404 | 0.0932 | Yes |
| 8 | Cdk2 | 1780 | 0.378 | 0.0990 | Yes |
| 9 | Ptpn11 | 1823 | 0.373 | 0.1144 | Yes |
| 10 | Trib3 | 1918 | 0.365 | 0.1269 | Yes |
| 11 | Actr2 | 2179 | 0.344 | 0.1300 | Yes |
| 12 | Cdkn1b | 2204 | 0.343 | 0.1449 | Yes |
| 13 | Mapk10 | 2253 | 0.340 | 0.1585 | Yes |
| 14 | Prkaa2 | 2308 | 0.335 | 0.1716 | Yes |
| 15 | Cab39 | 2341 | 0.333 | 0.1857 | Yes |
| 16 | Ube2n | 2509 | 0.321 | 0.1924 | Yes |
| 17 | Pikfyve | 2662 | 0.311 | 0.1993 | Yes |
| 18 | Arpc3 | 2937 | 0.292 | 0.1992 | No |
| 19 | Nck1 | 3739 | 0.245 | 0.1700 | No |
| 20 | Pitx2 | 3906 | 0.233 | 0.1726 | No |
| 21 | Calr | 4033 | 0.226 | 0.1768 | No |
| 22 | Prkar2a | 4199 | 0.219 | 0.1788 | No |
| 23 | Stat2 | 4293 | 0.214 | 0.1842 | No |
| 24 | Map2k3 | 4485 | 0.202 | 0.1840 | No |
| 25 | Pin1 | 4715 | 0.191 | 0.1814 | No |
| 26 | Mapk1 | 5311 | 0.163 | 0.1588 | No |
| 27 | Pten | 5324 | 0.162 | 0.1658 | No |
| 28 | Ube2d3 | 5327 | 0.162 | 0.1734 | No |
| 29 | Adcy2 | 5419 | 0.158 | 0.1762 | No |
| 30 | Il4 | 5462 | 0.155 | 0.1814 | No |
| 31 | Nod1 | 5635 | 0.147 | 0.1796 | No |
| 32 | Traf2 | 5694 | 0.144 | 0.1835 | No |
| 33 | Actr3 | 5757 | 0.142 | 0.1870 | No |
| 34 | Smad2 | 5857 | 0.139 | 0.1885 | No |
| 35 | Acaca | 6050 | 0.132 | 0.1850 | No |
| 36 | Ap2m1 | 6110 | 0.128 | 0.1881 | No |
| 37 | Map3k7 | 6354 | 0.118 | 0.1813 | No |
| 38 | Rac1 | 6511 | 0.111 | 0.1786 | No |
| 39 | Hras | 6585 | 0.108 | 0.1799 | No |
| 40 | Cdkn1a | 6987 | 0.092 | 0.1639 | No |
| 41 | Grb2 | 7016 | 0.090 | 0.1667 | No |
| 42 | Atf1 | 7039 | 0.089 | 0.1698 | No |
| 43 | Csnk2b | 7046 | 0.088 | 0.1737 | No |
| 44 | Mapkap1 | 7095 | 0.087 | 0.1753 | No |
| 45 | Eif4e | 7208 | 0.082 | 0.1735 | No |
| 46 | Gsk3b | 7903 | 0.053 | 0.1407 | No |
| 47 | Raf1 | 8295 | 0.036 | 0.1225 | No |
| 48 | Irak4 | 8296 | 0.036 | 0.1242 | No |
| 49 | Tbk1 | 8436 | 0.030 | 0.1186 | No |
| 50 | Ywhab | 8499 | 0.028 | 0.1167 | No |
| 51 | Them4 | 8747 | 0.018 | 0.1050 | No |
| 52 | Mknk2 | 8779 | 0.016 | 0.1042 | No |
| 53 | Ppp1ca | 8802 | 0.015 | 0.1038 | No |
| 54 | Pdk1 | 8869 | 0.012 | 0.1010 | No |
| 55 | Mapk8 | 9063 | 0.005 | 0.0914 | No |
| 56 | Il2rg | 9657 | -0.013 | 0.0619 | No |
| 57 | Sfn | 9895 | -0.022 | 0.0509 | No |
| 58 | Mapk9 | 10169 | -0.034 | 0.0387 | No |
| 59 | Cdk4 | 10287 | -0.039 | 0.0345 | No |
| 60 | Ripk1 | 10599 | -0.050 | 0.0211 | No |
| 61 | Sla | 10715 | -0.055 | 0.0179 | No |
| 62 | Dusp3 | 10845 | -0.061 | 0.0142 | No |
| 63 | Cfl1 | 10953 | -0.066 | 0.0119 | No |
| 64 | Ecsit | 11496 | -0.089 | -0.0115 | No |
| 65 | Prkag1 | 11558 | -0.092 | -0.0102 | No |
| 66 | Arhgdia | 11872 | -0.104 | -0.0212 | No |
| 67 | Sqstm1 | 11938 | -0.107 | -0.0195 | No |
| 68 | Itpr2 | 11939 | -0.107 | -0.0144 | No |
| 69 | Plcb1 | 12055 | -0.112 | -0.0150 | No |
| 70 | Lck | 12306 | -0.122 | -0.0219 | No |
| 71 | Vav3 | 12314 | -0.122 | -0.0165 | No |
| 72 | Pak4 | 12359 | -0.124 | -0.0129 | No |
| 73 | Grk2 | 12698 | -0.136 | -0.0236 | No |
| 74 | Cxcr4 | 12712 | -0.137 | -0.0178 | No |
| 75 | Pla2g12a | 12857 | -0.142 | -0.0184 | No |
| 76 | E2f1 | 12872 | -0.143 | -0.0124 | No |
| 77 | Rptor | 12982 | -0.147 | -0.0110 | No |
| 78 | Plcg1 | 13334 | -0.163 | -0.0212 | No |
| 79 | Tiam1 | 13471 | -0.169 | -0.0201 | No |
| 80 | Ppp2r1b | 13485 | -0.170 | -0.0128 | No |
| 81 | Ralb | 13496 | -0.170 | -0.0052 | No |
| 82 | Rit1 | 13866 | -0.185 | -0.0152 | No |
| 83 | Tsc2 | 14305 | -0.207 | -0.0277 | No |
| 84 | Pfn1 | 14384 | -0.210 | -0.0218 | No |
| 85 | Nfkbib | 14795 | -0.232 | -0.0317 | No |
| 86 | Cab39l | 14980 | -0.241 | -0.0297 | No |
| 87 | Egfr | 14999 | -0.242 | -0.0192 | No |
| 88 | Rps6ka1 | 15680 | -0.276 | -0.0407 | No |
| 89 | Mknk1 | 15792 | -0.283 | -0.0330 | No |
| 90 | Tnfrsf1a | 15836 | -0.286 | -0.0217 | No |
| 91 | Slc2a1 | 16284 | -0.313 | -0.0297 | No |
| 92 | Dapp1 | 16469 | -0.325 | -0.0237 | No |
| 93 | Ddit3 | 17097 | -0.365 | -0.0384 | No |
| 94 | Akt1s1 | 17299 | -0.381 | -0.0306 | No |
| 95 | Prkcb | 17543 | -0.401 | -0.0240 | No |
| 96 | Pik3r3 | 17807 | -0.424 | -0.0174 | No |
| 97 | Map2k6 | 18106 | -0.455 | -0.0111 | No |
| 98 | Camk4 | 19306 | -0.678 | -0.0401 | No |
| 99 | Gna14 | 19766 | -1.349 | 0.0003 | No |