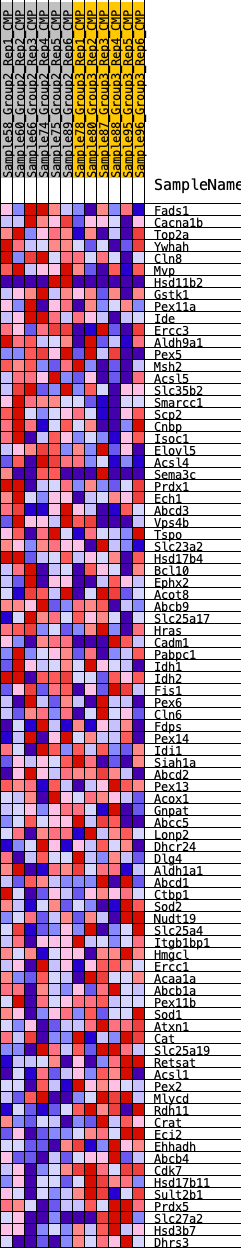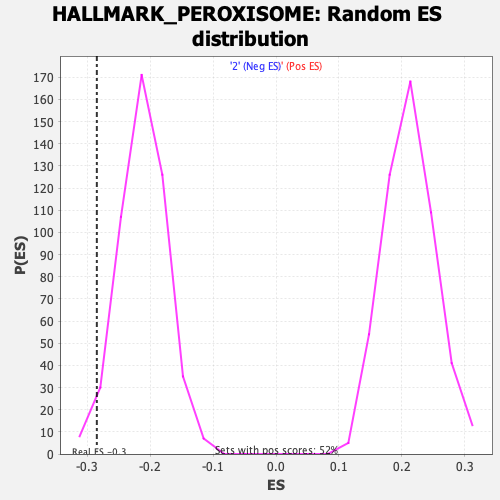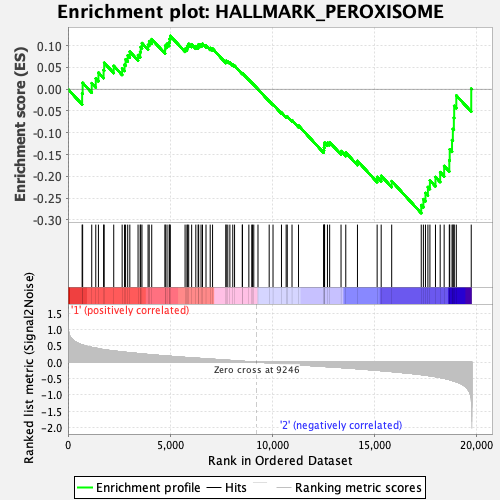
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group3.CMP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.28440702 |
| Normalized Enrichment Score (NES) | -1.3491862 |
| Nominal p-value | 0.02892562 |
| FDR q-value | 0.73700744 |
| FWER p-Value | 0.655 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fads1 | 692 | 0.528 | -0.0096 | No |
| 2 | Cacna1b | 717 | 0.522 | 0.0146 | No |
| 3 | Top2a | 1162 | 0.450 | 0.0138 | No |
| 4 | Ywhah | 1360 | 0.429 | 0.0246 | No |
| 5 | Cln8 | 1490 | 0.411 | 0.0380 | No |
| 6 | Mvp | 1742 | 0.381 | 0.0437 | No |
| 7 | Hsd11b2 | 1771 | 0.378 | 0.0606 | No |
| 8 | Gstk1 | 2239 | 0.341 | 0.0534 | No |
| 9 | Pex11a | 2651 | 0.312 | 0.0477 | No |
| 10 | Ide | 2768 | 0.303 | 0.0564 | No |
| 11 | Ercc3 | 2819 | 0.300 | 0.0684 | No |
| 12 | Aldh9a1 | 2923 | 0.293 | 0.0774 | No |
| 13 | Pex5 | 3027 | 0.285 | 0.0860 | No |
| 14 | Msh2 | 3430 | 0.262 | 0.0783 | No |
| 15 | Acsl5 | 3533 | 0.256 | 0.0856 | No |
| 16 | Slc35b2 | 3559 | 0.255 | 0.0967 | No |
| 17 | Smarcc1 | 3621 | 0.251 | 0.1057 | No |
| 18 | Scp2 | 3920 | 0.232 | 0.1019 | No |
| 19 | Cnbp | 3975 | 0.229 | 0.1102 | No |
| 20 | Isoc1 | 4099 | 0.223 | 0.1148 | No |
| 21 | Elovl5 | 4753 | 0.189 | 0.0908 | No |
| 22 | Acsl4 | 4757 | 0.189 | 0.0998 | No |
| 23 | Sema3c | 4848 | 0.185 | 0.1042 | No |
| 24 | Prdx1 | 4953 | 0.181 | 0.1077 | No |
| 25 | Ech1 | 4977 | 0.180 | 0.1153 | No |
| 26 | Abcd3 | 5009 | 0.178 | 0.1223 | No |
| 27 | Vps4b | 5732 | 0.143 | 0.0926 | No |
| 28 | Tspo | 5823 | 0.141 | 0.0949 | No |
| 29 | Slc23a2 | 5859 | 0.139 | 0.0998 | No |
| 30 | Hsd17b4 | 5910 | 0.137 | 0.1040 | No |
| 31 | Bcl10 | 6054 | 0.132 | 0.1031 | No |
| 32 | Ephx2 | 6258 | 0.122 | 0.0987 | No |
| 33 | Acot8 | 6370 | 0.117 | 0.0987 | No |
| 34 | Abcb9 | 6397 | 0.116 | 0.1030 | No |
| 35 | Slc25a17 | 6514 | 0.111 | 0.1025 | No |
| 36 | Hras | 6585 | 0.108 | 0.1042 | No |
| 37 | Cadm1 | 6757 | 0.102 | 0.1004 | No |
| 38 | Pabpc1 | 6962 | 0.093 | 0.0946 | No |
| 39 | Idh1 | 7078 | 0.087 | 0.0930 | No |
| 40 | Idh2 | 7730 | 0.060 | 0.0628 | No |
| 41 | Fis1 | 7742 | 0.059 | 0.0651 | No |
| 42 | Pex6 | 7823 | 0.056 | 0.0637 | No |
| 43 | Cln6 | 7926 | 0.052 | 0.0611 | No |
| 44 | Fdps | 8067 | 0.046 | 0.0562 | No |
| 45 | Pex14 | 8155 | 0.042 | 0.0538 | No |
| 46 | Idi1 | 8533 | 0.026 | 0.0359 | No |
| 47 | Siah1a | 8542 | 0.026 | 0.0367 | No |
| 48 | Abcd2 | 8855 | 0.013 | 0.0215 | No |
| 49 | Pex13 | 8997 | 0.008 | 0.0147 | No |
| 50 | Acox1 | 9036 | 0.006 | 0.0131 | No |
| 51 | Gnpat | 9096 | 0.004 | 0.0103 | No |
| 52 | Abcc5 | 9302 | -0.001 | -0.0001 | No |
| 53 | Lonp2 | 9850 | -0.020 | -0.0269 | No |
| 54 | Dhcr24 | 10038 | -0.029 | -0.0350 | No |
| 55 | Dlg4 | 10452 | -0.044 | -0.0538 | No |
| 56 | Aldh1a1 | 10681 | -0.054 | -0.0627 | No |
| 57 | Abcd1 | 10735 | -0.056 | -0.0627 | No |
| 58 | Ctbp1 | 10968 | -0.067 | -0.0712 | No |
| 59 | Sod2 | 11286 | -0.080 | -0.0835 | No |
| 60 | Nudt19 | 12516 | -0.130 | -0.1396 | No |
| 61 | Slc25a4 | 12535 | -0.131 | -0.1341 | No |
| 62 | Itgb1bp1 | 12552 | -0.132 | -0.1285 | No |
| 63 | Hmgcl | 12563 | -0.132 | -0.1226 | No |
| 64 | Ercc1 | 12703 | -0.137 | -0.1231 | No |
| 65 | Acaa1a | 12811 | -0.140 | -0.1217 | No |
| 66 | Abcb1a | 13368 | -0.164 | -0.1420 | No |
| 67 | Pex11b | 13600 | -0.175 | -0.1453 | No |
| 68 | Sod1 | 14172 | -0.201 | -0.1645 | No |
| 69 | Atxn1 | 15136 | -0.248 | -0.2014 | No |
| 70 | Cat | 15334 | -0.259 | -0.1988 | No |
| 71 | Slc25a19 | 15848 | -0.287 | -0.2110 | No |
| 72 | Retsat | 17295 | -0.380 | -0.2660 | Yes |
| 73 | Acsl1 | 17400 | -0.389 | -0.2524 | Yes |
| 74 | Pex2 | 17502 | -0.398 | -0.2382 | Yes |
| 75 | Mlycd | 17621 | -0.407 | -0.2244 | Yes |
| 76 | Rdh11 | 17720 | -0.416 | -0.2092 | Yes |
| 77 | Crat | 17992 | -0.443 | -0.2015 | Yes |
| 78 | Eci2 | 18222 | -0.469 | -0.1903 | Yes |
| 79 | Ehhadh | 18417 | -0.495 | -0.1762 | Yes |
| 80 | Abcb4 | 18669 | -0.533 | -0.1631 | Yes |
| 81 | Cdk7 | 18690 | -0.536 | -0.1381 | Yes |
| 82 | Hsd17b11 | 18803 | -0.559 | -0.1167 | Yes |
| 83 | Sult2b1 | 18843 | -0.566 | -0.0912 | Yes |
| 84 | Prdx5 | 18892 | -0.577 | -0.0657 | Yes |
| 85 | Slc27a2 | 18908 | -0.579 | -0.0384 | Yes |
| 86 | Hsd3b7 | 19012 | -0.600 | -0.0145 | Yes |
| 87 | Dhrs3 | 19743 | -1.093 | 0.0014 | Yes |