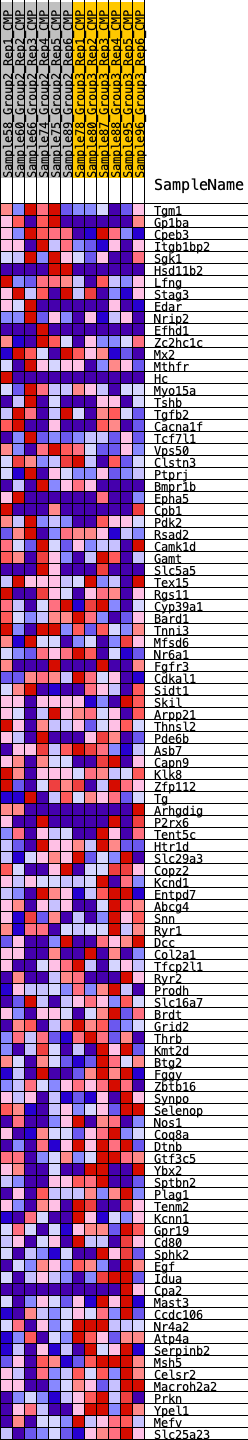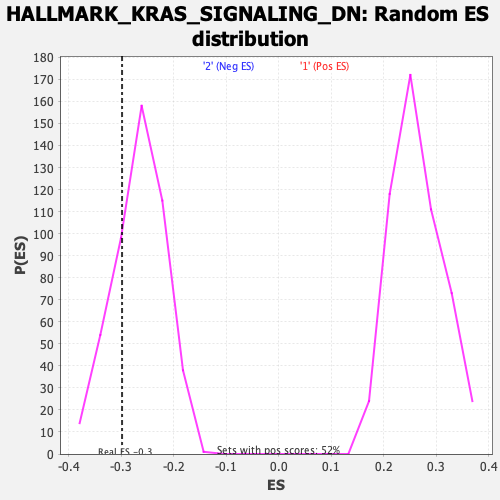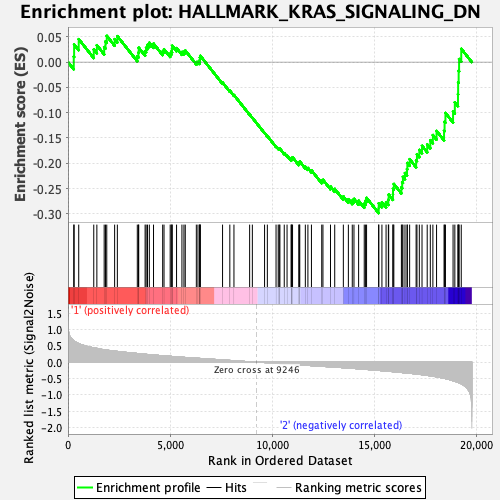
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group3.CMP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.29850546 |
| Normalized Enrichment Score (NES) | -1.1292491 |
| Nominal p-value | 0.24686192 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.938 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tgm1 | 282 | 0.655 | 0.0109 | No |
| 2 | Gp1ba | 300 | 0.648 | 0.0349 | No |
| 3 | Cpeb3 | 525 | 0.566 | 0.0453 | No |
| 4 | Itgb1bp2 | 1261 | 0.439 | 0.0249 | No |
| 5 | Sgk1 | 1413 | 0.422 | 0.0334 | No |
| 6 | Hsd11b2 | 1771 | 0.378 | 0.0298 | No |
| 7 | Lfng | 1835 | 0.372 | 0.0409 | No |
| 8 | Stag3 | 1898 | 0.367 | 0.0519 | No |
| 9 | Edar | 2283 | 0.338 | 0.0454 | No |
| 10 | Nrip2 | 2416 | 0.328 | 0.0513 | No |
| 11 | Efhd1 | 3393 | 0.264 | 0.0118 | No |
| 12 | Zc2hc1c | 3451 | 0.261 | 0.0189 | No |
| 13 | Mx2 | 3463 | 0.260 | 0.0284 | No |
| 14 | Mthfr | 3775 | 0.242 | 0.0219 | No |
| 15 | Hc | 3825 | 0.238 | 0.0285 | No |
| 16 | Myo15a | 3892 | 0.234 | 0.0342 | No |
| 17 | Tshb | 3982 | 0.229 | 0.0385 | No |
| 18 | Tgfb2 | 4187 | 0.220 | 0.0365 | No |
| 19 | Cacna1f | 4635 | 0.195 | 0.0213 | No |
| 20 | Tcf7l1 | 4701 | 0.192 | 0.0254 | No |
| 21 | Vps50 | 5003 | 0.178 | 0.0170 | No |
| 22 | Clstn3 | 5056 | 0.176 | 0.0211 | No |
| 23 | Ptprj | 5094 | 0.174 | 0.0259 | No |
| 24 | Bmpr1b | 5097 | 0.173 | 0.0325 | No |
| 25 | Epha5 | 5315 | 0.163 | 0.0277 | No |
| 26 | Cpb1 | 5578 | 0.149 | 0.0201 | No |
| 27 | Pdk2 | 5677 | 0.145 | 0.0207 | No |
| 28 | Rsad2 | 5742 | 0.143 | 0.0229 | No |
| 29 | Camk1d | 6283 | 0.121 | 0.0001 | No |
| 30 | Gamt | 6363 | 0.117 | 0.0006 | No |
| 31 | Slc5a5 | 6450 | 0.113 | 0.0006 | No |
| 32 | Tex15 | 6456 | 0.113 | 0.0047 | No |
| 33 | Rgs11 | 6459 | 0.113 | 0.0089 | No |
| 34 | Cyp39a1 | 6476 | 0.112 | 0.0124 | No |
| 35 | Bard1 | 7567 | 0.066 | -0.0404 | No |
| 36 | Tnni3 | 7923 | 0.052 | -0.0565 | No |
| 37 | Mfsd6 | 8125 | 0.043 | -0.0651 | No |
| 38 | Nr6a1 | 8893 | 0.011 | -0.1036 | No |
| 39 | Fgfr3 | 9026 | 0.007 | -0.1101 | No |
| 40 | Cdkal1 | 9624 | -0.012 | -0.1400 | No |
| 41 | Sidt1 | 9760 | -0.017 | -0.1462 | No |
| 42 | Skil | 10186 | -0.035 | -0.1665 | No |
| 43 | Arpp21 | 10292 | -0.039 | -0.1703 | No |
| 44 | Thnsl2 | 10342 | -0.041 | -0.1712 | No |
| 45 | Pde6b | 10375 | -0.042 | -0.1712 | No |
| 46 | Asb7 | 10588 | -0.050 | -0.1801 | No |
| 47 | Capn9 | 10725 | -0.056 | -0.1849 | No |
| 48 | Klk8 | 10927 | -0.065 | -0.1926 | No |
| 49 | Zfp112 | 10935 | -0.065 | -0.1904 | No |
| 50 | Tg | 10962 | -0.066 | -0.1892 | No |
| 51 | Arhgdig | 10994 | -0.068 | -0.1882 | No |
| 52 | P2rx6 | 11300 | -0.081 | -0.2006 | No |
| 53 | Tent5c | 11328 | -0.082 | -0.1988 | No |
| 54 | Htr1d | 11348 | -0.083 | -0.1965 | No |
| 55 | Slc29a3 | 11620 | -0.094 | -0.2067 | No |
| 56 | Copz2 | 11746 | -0.099 | -0.2093 | No |
| 57 | Kcnd1 | 11919 | -0.106 | -0.2139 | No |
| 58 | Entpd7 | 12423 | -0.127 | -0.2346 | No |
| 59 | Abcg4 | 12482 | -0.129 | -0.2326 | No |
| 60 | Snn | 12852 | -0.142 | -0.2459 | No |
| 61 | Ryr1 | 13059 | -0.150 | -0.2506 | No |
| 62 | Dcc | 13481 | -0.169 | -0.2655 | No |
| 63 | Col2a1 | 13727 | -0.180 | -0.2710 | No |
| 64 | Tfcp2l1 | 13918 | -0.188 | -0.2735 | No |
| 65 | Ryr2 | 14001 | -0.191 | -0.2703 | No |
| 66 | Prodh | 14229 | -0.204 | -0.2740 | No |
| 67 | Slc16a7 | 14504 | -0.216 | -0.2796 | No |
| 68 | Brdt | 14562 | -0.219 | -0.2741 | No |
| 69 | Grid2 | 14618 | -0.222 | -0.2684 | No |
| 70 | Thrb | 15212 | -0.253 | -0.2888 | Yes |
| 71 | Kmt2d | 15214 | -0.253 | -0.2791 | Yes |
| 72 | Btg2 | 15370 | -0.261 | -0.2769 | Yes |
| 73 | Fggy | 15573 | -0.271 | -0.2768 | Yes |
| 74 | Zbtb16 | 15685 | -0.277 | -0.2718 | Yes |
| 75 | Synpo | 15701 | -0.278 | -0.2619 | Yes |
| 76 | Selenop | 15909 | -0.291 | -0.2612 | Yes |
| 77 | Nos1 | 15913 | -0.291 | -0.2502 | Yes |
| 78 | Coq8a | 15953 | -0.293 | -0.2409 | Yes |
| 79 | Dtnb | 16323 | -0.315 | -0.2475 | Yes |
| 80 | Gtf3c5 | 16362 | -0.318 | -0.2372 | Yes |
| 81 | Ybx2 | 16399 | -0.320 | -0.2267 | Yes |
| 82 | Sptbn2 | 16499 | -0.327 | -0.2192 | Yes |
| 83 | Plag1 | 16599 | -0.332 | -0.2114 | Yes |
| 84 | Tenm2 | 16615 | -0.334 | -0.1993 | Yes |
| 85 | Kcnn1 | 16727 | -0.341 | -0.1919 | Yes |
| 86 | Gpr19 | 17044 | -0.361 | -0.1940 | Yes |
| 87 | Cd80 | 17085 | -0.364 | -0.1821 | Yes |
| 88 | Sphk2 | 17201 | -0.374 | -0.1736 | Yes |
| 89 | Egf | 17333 | -0.384 | -0.1654 | Yes |
| 90 | Idua | 17587 | -0.405 | -0.1627 | Yes |
| 91 | Cpa2 | 17739 | -0.418 | -0.1543 | Yes |
| 92 | Mast3 | 17865 | -0.430 | -0.1442 | Yes |
| 93 | Ccdc106 | 18045 | -0.447 | -0.1361 | Yes |
| 94 | Nr4a2 | 18412 | -0.495 | -0.1356 | Yes |
| 95 | Atp4a | 18438 | -0.497 | -0.1178 | Yes |
| 96 | Serpinb2 | 18480 | -0.503 | -0.1005 | Yes |
| 97 | Msh5 | 18853 | -0.568 | -0.0975 | Yes |
| 98 | Celsr2 | 18940 | -0.585 | -0.0794 | Yes |
| 99 | Macroh2a2 | 19094 | -0.618 | -0.0634 | Yes |
| 100 | Prkn | 19102 | -0.619 | -0.0399 | Yes |
| 101 | Ypel1 | 19127 | -0.625 | -0.0171 | Yes |
| 102 | Mefv | 19149 | -0.631 | 0.0061 | Yes |
| 103 | Slc25a23 | 19254 | -0.662 | 0.0263 | Yes |