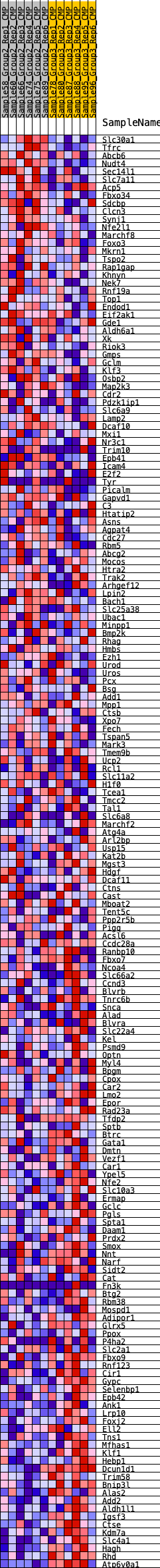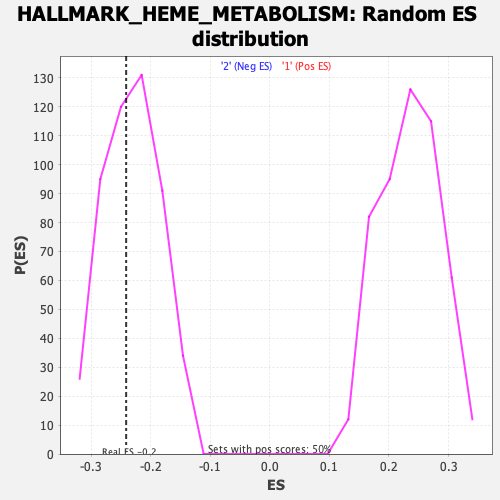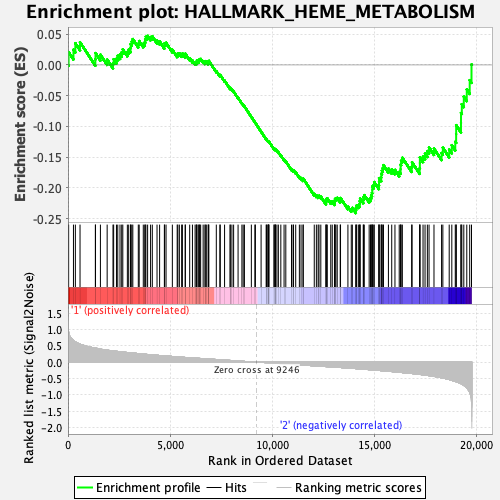
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group3.CMP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_HEME_METABOLISM |
| Enrichment Score (ES) | -0.24107903 |
| Normalized Enrichment Score (NES) | -1.0442796 |
| Nominal p-value | 0.43259558 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.982 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Slc30a1 | 31 | 0.926 | 0.0209 | No |
| 2 | Tfrc | 267 | 0.662 | 0.0249 | No |
| 3 | Abcb6 | 360 | 0.623 | 0.0353 | No |
| 4 | Nudt4 | 589 | 0.548 | 0.0370 | No |
| 5 | Sec14l1 | 1339 | 0.431 | 0.0092 | No |
| 6 | Slc7a11 | 1347 | 0.430 | 0.0193 | No |
| 7 | Acp5 | 1587 | 0.398 | 0.0167 | No |
| 8 | Fbxo34 | 1917 | 0.366 | 0.0088 | No |
| 9 | Sdcbp | 2203 | 0.343 | 0.0026 | No |
| 10 | Clcn3 | 2235 | 0.341 | 0.0092 | No |
| 11 | Synj1 | 2373 | 0.331 | 0.0103 | No |
| 12 | Nfe2l1 | 2436 | 0.326 | 0.0150 | No |
| 13 | Marchf8 | 2546 | 0.319 | 0.0172 | No |
| 14 | Foxo3 | 2630 | 0.314 | 0.0206 | No |
| 15 | Mkrn1 | 2683 | 0.309 | 0.0254 | No |
| 16 | Tspo2 | 2906 | 0.295 | 0.0212 | No |
| 17 | Rap1gap | 2971 | 0.290 | 0.0250 | No |
| 18 | Khnyn | 3061 | 0.284 | 0.0273 | No |
| 19 | Nek7 | 3068 | 0.283 | 0.0339 | No |
| 20 | Rnf19a | 3122 | 0.280 | 0.0380 | No |
| 21 | Top1 | 3173 | 0.278 | 0.0421 | No |
| 22 | Endod1 | 3442 | 0.262 | 0.0348 | No |
| 23 | Eif2ak1 | 3485 | 0.259 | 0.0389 | No |
| 24 | Gde1 | 3686 | 0.247 | 0.0347 | No |
| 25 | Aldh6a1 | 3760 | 0.243 | 0.0369 | No |
| 26 | Xk | 3780 | 0.242 | 0.0418 | No |
| 27 | Riok3 | 3813 | 0.239 | 0.0459 | No |
| 28 | Gmps | 3898 | 0.233 | 0.0473 | No |
| 29 | Gclm | 4043 | 0.226 | 0.0454 | No |
| 30 | Klf3 | 4128 | 0.222 | 0.0465 | No |
| 31 | Osbp2 | 4359 | 0.209 | 0.0399 | No |
| 32 | Map2k3 | 4485 | 0.202 | 0.0384 | No |
| 33 | Cdr2 | 4718 | 0.191 | 0.0312 | No |
| 34 | Pdzk1ip1 | 4729 | 0.190 | 0.0353 | No |
| 35 | Slc6a9 | 4800 | 0.188 | 0.0362 | No |
| 36 | Lamp2 | 5109 | 0.173 | 0.0247 | No |
| 37 | Dcaf10 | 5346 | 0.161 | 0.0166 | No |
| 38 | Mxi1 | 5378 | 0.160 | 0.0189 | No |
| 39 | Nr3c1 | 5452 | 0.156 | 0.0189 | No |
| 40 | Trim10 | 5565 | 0.150 | 0.0168 | No |
| 41 | Epb41 | 5601 | 0.149 | 0.0187 | No |
| 42 | Icam4 | 5741 | 0.143 | 0.0150 | No |
| 43 | E2f2 | 5743 | 0.143 | 0.0184 | No |
| 44 | Tyr | 5958 | 0.135 | 0.0108 | No |
| 45 | Picalm | 6093 | 0.129 | 0.0071 | No |
| 46 | Gapvd1 | 6223 | 0.123 | 0.0035 | No |
| 47 | C3 | 6282 | 0.121 | 0.0034 | No |
| 48 | Htatip2 | 6300 | 0.120 | 0.0055 | No |
| 49 | Asns | 6317 | 0.119 | 0.0076 | No |
| 50 | Agpat4 | 6401 | 0.115 | 0.0061 | No |
| 51 | Cdc27 | 6417 | 0.115 | 0.0081 | No |
| 52 | Rbm5 | 6446 | 0.113 | 0.0095 | No |
| 53 | Abcg2 | 6494 | 0.112 | 0.0098 | No |
| 54 | Mocos | 6617 | 0.106 | 0.0061 | No |
| 55 | Htra2 | 6711 | 0.103 | 0.0039 | No |
| 56 | Trak2 | 6716 | 0.103 | 0.0062 | No |
| 57 | Arhgef12 | 6772 | 0.101 | 0.0058 | No |
| 58 | Lpin2 | 6867 | 0.098 | 0.0034 | No |
| 59 | Bach1 | 6868 | 0.097 | 0.0057 | No |
| 60 | Slc25a38 | 6896 | 0.096 | 0.0067 | No |
| 61 | Ubac1 | 7261 | 0.079 | -0.0100 | No |
| 62 | Minpp1 | 7436 | 0.071 | -0.0171 | No |
| 63 | Bmp2k | 7454 | 0.070 | -0.0163 | No |
| 64 | Rhag | 7666 | 0.062 | -0.0256 | No |
| 65 | Hmbs | 7936 | 0.051 | -0.0380 | No |
| 66 | Ezh1 | 7944 | 0.051 | -0.0372 | No |
| 67 | Urod | 8026 | 0.048 | -0.0401 | No |
| 68 | Uros | 8109 | 0.043 | -0.0433 | No |
| 69 | Pcx | 8327 | 0.034 | -0.0535 | No |
| 70 | Bsg | 8515 | 0.027 | -0.0624 | No |
| 71 | Add1 | 8577 | 0.024 | -0.0649 | No |
| 72 | Mpp1 | 8638 | 0.021 | -0.0675 | No |
| 73 | Ctsb | 8977 | 0.009 | -0.0845 | No |
| 74 | Xpo7 | 9150 | 0.003 | -0.0932 | No |
| 75 | Fech | 9175 | 0.002 | -0.0944 | No |
| 76 | Tspan5 | 9454 | -0.005 | -0.1085 | No |
| 77 | Mark3 | 9702 | -0.015 | -0.1207 | No |
| 78 | Tmem9b | 9721 | -0.015 | -0.1213 | No |
| 79 | Ucp2 | 9790 | -0.018 | -0.1243 | No |
| 80 | Rcl1 | 9795 | -0.018 | -0.1241 | No |
| 81 | Slc11a2 | 9814 | -0.019 | -0.1245 | No |
| 82 | H1f0 | 9821 | -0.019 | -0.1244 | No |
| 83 | Tcea1 | 9843 | -0.020 | -0.1250 | No |
| 84 | Tmcc2 | 10086 | -0.031 | -0.1366 | No |
| 85 | Tal1 | 10096 | -0.031 | -0.1363 | No |
| 86 | Slc6a8 | 10143 | -0.033 | -0.1378 | No |
| 87 | Marchf2 | 10150 | -0.033 | -0.1373 | No |
| 88 | Atg4a | 10156 | -0.033 | -0.1368 | No |
| 89 | Arl2bp | 10228 | -0.036 | -0.1395 | No |
| 90 | Usp15 | 10299 | -0.039 | -0.1421 | No |
| 91 | Kat2b | 10420 | -0.044 | -0.1472 | No |
| 92 | Mgst3 | 10579 | -0.050 | -0.1541 | No |
| 93 | Hdgf | 10664 | -0.054 | -0.1570 | No |
| 94 | Dcaf11 | 10943 | -0.065 | -0.1696 | No |
| 95 | Ctns | 11018 | -0.069 | -0.1717 | No |
| 96 | Cast | 11042 | -0.070 | -0.1712 | No |
| 97 | Mboat2 | 11149 | -0.075 | -0.1748 | No |
| 98 | Tent5c | 11328 | -0.082 | -0.1819 | No |
| 99 | Ppp2r5b | 11410 | -0.086 | -0.1839 | No |
| 100 | Pigq | 11507 | -0.090 | -0.1867 | No |
| 101 | Acsl6 | 11518 | -0.090 | -0.1850 | No |
| 102 | Ccdc28a | 12057 | -0.112 | -0.2097 | No |
| 103 | Ranbp10 | 12160 | -0.116 | -0.2121 | No |
| 104 | Fbxo7 | 12246 | -0.120 | -0.2135 | No |
| 105 | Ncoa4 | 12288 | -0.121 | -0.2127 | No |
| 106 | Slc66a2 | 12378 | -0.125 | -0.2142 | No |
| 107 | Ccnd3 | 12630 | -0.134 | -0.2238 | No |
| 108 | Blvrb | 12639 | -0.135 | -0.2209 | No |
| 109 | Tnrc6b | 12644 | -0.135 | -0.2178 | No |
| 110 | Snca | 12695 | -0.136 | -0.2171 | No |
| 111 | Alad | 12865 | -0.142 | -0.2223 | No |
| 112 | Blvra | 12937 | -0.146 | -0.2224 | No |
| 113 | Slc22a4 | 13047 | -0.150 | -0.2243 | No |
| 114 | Kel | 13057 | -0.150 | -0.2211 | No |
| 115 | Psmd9 | 13074 | -0.150 | -0.2183 | No |
| 116 | Optn | 13115 | -0.152 | -0.2166 | No |
| 117 | Myl4 | 13181 | -0.156 | -0.2162 | No |
| 118 | Bpgm | 13328 | -0.163 | -0.2197 | No |
| 119 | Cpox | 13343 | -0.163 | -0.2164 | No |
| 120 | Car2 | 13705 | -0.179 | -0.2305 | No |
| 121 | Lmo2 | 13869 | -0.185 | -0.2344 | No |
| 122 | Epor | 13922 | -0.188 | -0.2325 | No |
| 123 | Rad23a | 14092 | -0.196 | -0.2363 | Yes |
| 124 | Tfdp2 | 14103 | -0.197 | -0.2320 | Yes |
| 125 | Sptb | 14131 | -0.199 | -0.2286 | Yes |
| 126 | Btrc | 14216 | -0.203 | -0.2280 | Yes |
| 127 | Gata1 | 14280 | -0.206 | -0.2262 | Yes |
| 128 | Dmtn | 14287 | -0.206 | -0.2215 | Yes |
| 129 | Vezf1 | 14303 | -0.207 | -0.2172 | Yes |
| 130 | Car1 | 14456 | -0.214 | -0.2198 | Yes |
| 131 | Ypel5 | 14466 | -0.214 | -0.2151 | Yes |
| 132 | Nfe2 | 14506 | -0.216 | -0.2118 | Yes |
| 133 | Slc10a3 | 14745 | -0.229 | -0.2184 | Yes |
| 134 | Ermap | 14812 | -0.233 | -0.2161 | Yes |
| 135 | Gclc | 14844 | -0.234 | -0.2120 | Yes |
| 136 | Pgls | 14883 | -0.236 | -0.2082 | Yes |
| 137 | Spta1 | 14899 | -0.237 | -0.2033 | Yes |
| 138 | Daam1 | 14900 | -0.237 | -0.1975 | Yes |
| 139 | Prdx2 | 14971 | -0.240 | -0.1953 | Yes |
| 140 | Smox | 14993 | -0.241 | -0.1905 | Yes |
| 141 | Nnt | 15222 | -0.253 | -0.1960 | Yes |
| 142 | Narf | 15229 | -0.254 | -0.1901 | Yes |
| 143 | Sidt2 | 15232 | -0.254 | -0.1841 | Yes |
| 144 | Cat | 15334 | -0.259 | -0.1829 | Yes |
| 145 | Fn3k | 15339 | -0.259 | -0.1769 | Yes |
| 146 | Btg2 | 15370 | -0.261 | -0.1721 | Yes |
| 147 | Rbm38 | 15414 | -0.263 | -0.1679 | Yes |
| 148 | Mospd1 | 15446 | -0.264 | -0.1630 | Yes |
| 149 | Adipor1 | 15688 | -0.277 | -0.1686 | Yes |
| 150 | Glrx5 | 15850 | -0.287 | -0.1699 | Yes |
| 151 | Ppox | 16008 | -0.297 | -0.1707 | Yes |
| 152 | P4ha2 | 16217 | -0.309 | -0.1738 | Yes |
| 153 | Slc2a1 | 16284 | -0.313 | -0.1696 | Yes |
| 154 | Fbxo9 | 16286 | -0.313 | -0.1621 | Yes |
| 155 | Rnf123 | 16310 | -0.315 | -0.1556 | Yes |
| 156 | Cir1 | 16370 | -0.319 | -0.1509 | Yes |
| 157 | Gypc | 16826 | -0.346 | -0.1657 | Yes |
| 158 | Selenbp1 | 16846 | -0.347 | -0.1583 | Yes |
| 159 | Epb42 | 17219 | -0.375 | -0.1682 | Yes |
| 160 | Ank1 | 17228 | -0.375 | -0.1595 | Yes |
| 161 | Lrp10 | 17229 | -0.375 | -0.1504 | Yes |
| 162 | Foxj2 | 17383 | -0.388 | -0.1488 | Yes |
| 163 | Ell2 | 17481 | -0.396 | -0.1441 | Yes |
| 164 | Tns1 | 17595 | -0.405 | -0.1401 | Yes |
| 165 | Mfhas1 | 17677 | -0.413 | -0.1342 | Yes |
| 166 | Klf1 | 17919 | -0.435 | -0.1359 | Yes |
| 167 | Hebp1 | 18290 | -0.478 | -0.1432 | Yes |
| 168 | Dcun1d1 | 18353 | -0.487 | -0.1346 | Yes |
| 169 | Trim58 | 18664 | -0.532 | -0.1375 | Yes |
| 170 | Bnip3l | 18792 | -0.557 | -0.1305 | Yes |
| 171 | Alas2 | 18961 | -0.589 | -0.1248 | Yes |
| 172 | Add2 | 19004 | -0.598 | -0.1124 | Yes |
| 173 | Aldh1l1 | 19006 | -0.598 | -0.0980 | Yes |
| 174 | Igsf3 | 19241 | -0.657 | -0.0940 | Yes |
| 175 | Ctse | 19243 | -0.659 | -0.0781 | Yes |
| 176 | Kdm7a | 19282 | -0.670 | -0.0638 | Yes |
| 177 | Slc4a1 | 19379 | -0.707 | -0.0516 | Yes |
| 178 | Hagh | 19525 | -0.785 | -0.0399 | Yes |
| 179 | Rhd | 19668 | -0.924 | -0.0248 | Yes |
| 180 | Atp6v0a1 | 19756 | -1.236 | 0.0008 | Yes |