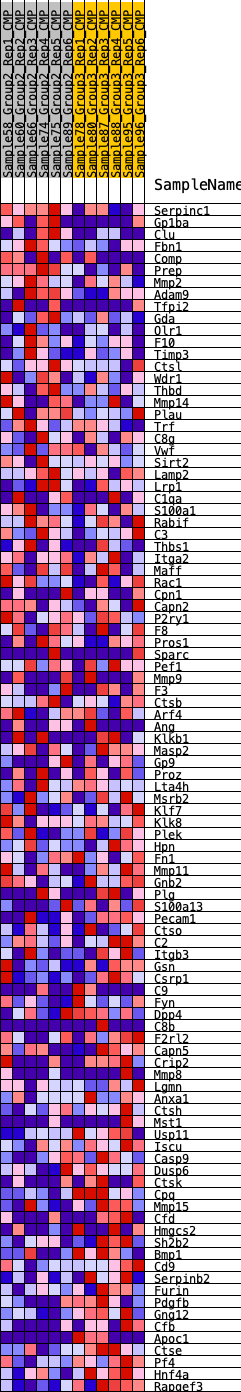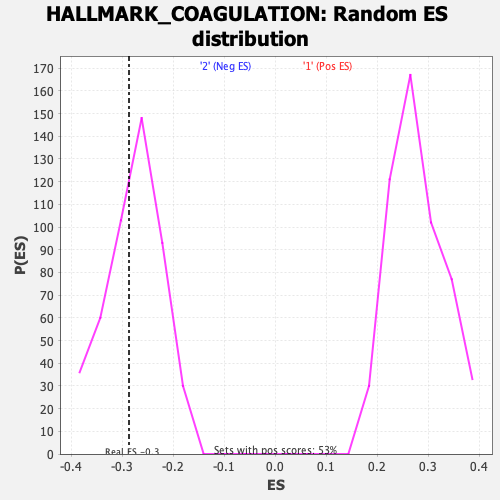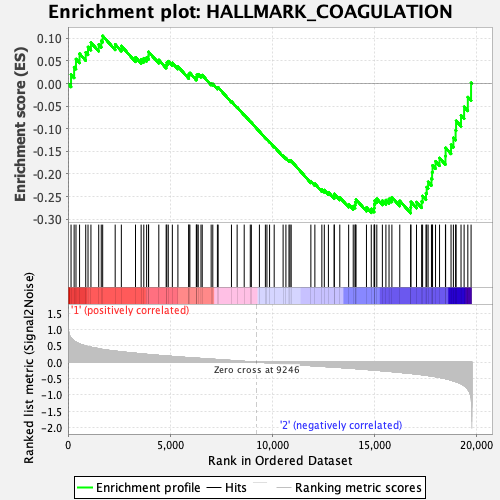
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group3.CMP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.2865793 |
| Normalized Enrichment Score (NES) | -1.0315757 |
| Nominal p-value | 0.41276595 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.983 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Serpinc1 | 149 | 0.744 | 0.0200 | No |
| 2 | Gp1ba | 300 | 0.648 | 0.0364 | No |
| 3 | Clu | 391 | 0.608 | 0.0544 | No |
| 4 | Fbn1 | 566 | 0.556 | 0.0661 | No |
| 5 | Comp | 868 | 0.492 | 0.0690 | No |
| 6 | Prep | 980 | 0.474 | 0.0810 | No |
| 7 | Mmp2 | 1123 | 0.454 | 0.0906 | No |
| 8 | Adam9 | 1504 | 0.408 | 0.0864 | No |
| 9 | Tfpi2 | 1628 | 0.393 | 0.0947 | No |
| 10 | Gda | 1698 | 0.386 | 0.1055 | No |
| 11 | Olr1 | 2310 | 0.335 | 0.0868 | No |
| 12 | F10 | 2612 | 0.315 | 0.0832 | No |
| 13 | Timp3 | 3305 | 0.270 | 0.0580 | No |
| 14 | Ctsl | 3581 | 0.253 | 0.0534 | No |
| 15 | Wdr1 | 3711 | 0.246 | 0.0560 | No |
| 16 | Thbd | 3850 | 0.236 | 0.0577 | No |
| 17 | Mmp14 | 3944 | 0.231 | 0.0616 | No |
| 18 | Plau | 3945 | 0.231 | 0.0701 | No |
| 19 | Trf | 4446 | 0.204 | 0.0523 | No |
| 20 | C8g | 4806 | 0.187 | 0.0410 | No |
| 21 | Vwf | 4837 | 0.186 | 0.0463 | No |
| 22 | Sirt2 | 4909 | 0.183 | 0.0495 | No |
| 23 | Lamp2 | 5109 | 0.173 | 0.0458 | No |
| 24 | Lrp1 | 5381 | 0.160 | 0.0379 | No |
| 25 | C1qa | 5913 | 0.137 | 0.0160 | No |
| 26 | S100a1 | 5917 | 0.137 | 0.0210 | No |
| 27 | Rabif | 5967 | 0.135 | 0.0235 | No |
| 28 | C3 | 6282 | 0.121 | 0.0120 | No |
| 29 | Thbs1 | 6304 | 0.120 | 0.0154 | No |
| 30 | Itga2 | 6312 | 0.120 | 0.0194 | No |
| 31 | Maff | 6376 | 0.117 | 0.0206 | No |
| 32 | Rac1 | 6511 | 0.111 | 0.0179 | No |
| 33 | Cpn1 | 6572 | 0.108 | 0.0188 | No |
| 34 | Capn2 | 7009 | 0.090 | 0.0000 | No |
| 35 | P2ry1 | 7089 | 0.087 | -0.0008 | No |
| 36 | F8 | 7315 | 0.077 | -0.0094 | No |
| 37 | Pros1 | 7351 | 0.075 | -0.0084 | No |
| 38 | Sparc | 8003 | 0.049 | -0.0397 | No |
| 39 | Pef1 | 8281 | 0.036 | -0.0524 | No |
| 40 | Mmp9 | 8627 | 0.022 | -0.0691 | No |
| 41 | F3 | 8925 | 0.011 | -0.0838 | No |
| 42 | Ctsb | 8977 | 0.009 | -0.0861 | No |
| 43 | Arf4 | 9372 | -0.003 | -0.1060 | No |
| 44 | Ang | 9664 | -0.013 | -0.1203 | No |
| 45 | Klkb1 | 9740 | -0.016 | -0.1235 | No |
| 46 | Masp2 | 9869 | -0.021 | -0.1292 | No |
| 47 | Gp9 | 10095 | -0.031 | -0.1395 | No |
| 48 | Proz | 10532 | -0.048 | -0.1599 | No |
| 49 | Lta4h | 10669 | -0.054 | -0.1648 | No |
| 50 | Msrb2 | 10820 | -0.060 | -0.1702 | No |
| 51 | Klf7 | 10869 | -0.062 | -0.1704 | No |
| 52 | Klk8 | 10927 | -0.065 | -0.1709 | No |
| 53 | Plek | 11891 | -0.105 | -0.2159 | No |
| 54 | Hpn | 12087 | -0.113 | -0.2216 | No |
| 55 | Fn1 | 12429 | -0.127 | -0.2343 | No |
| 56 | Mmp11 | 12549 | -0.131 | -0.2355 | No |
| 57 | Gnb2 | 12750 | -0.138 | -0.2405 | No |
| 58 | Plg | 13032 | -0.149 | -0.2492 | No |
| 59 | S100a13 | 13038 | -0.149 | -0.2440 | No |
| 60 | Pecam1 | 13307 | -0.162 | -0.2516 | No |
| 61 | Ctso | 13741 | -0.180 | -0.2669 | No |
| 62 | C2 | 13965 | -0.190 | -0.2712 | No |
| 63 | Itgb3 | 14054 | -0.194 | -0.2685 | No |
| 64 | Gsn | 14066 | -0.195 | -0.2618 | No |
| 65 | Csrp1 | 14102 | -0.197 | -0.2563 | No |
| 66 | C9 | 14613 | -0.222 | -0.2740 | No |
| 67 | Fyn | 14848 | -0.234 | -0.2772 | Yes |
| 68 | Dpp4 | 14988 | -0.241 | -0.2754 | Yes |
| 69 | C8b | 15000 | -0.242 | -0.2670 | Yes |
| 70 | F2rl2 | 15017 | -0.242 | -0.2588 | Yes |
| 71 | Capn5 | 15120 | -0.247 | -0.2548 | Yes |
| 72 | Crip2 | 15392 | -0.262 | -0.2589 | Yes |
| 73 | Mmp8 | 15565 | -0.270 | -0.2576 | Yes |
| 74 | Lgmn | 15718 | -0.279 | -0.2550 | Yes |
| 75 | Anxa1 | 15858 | -0.288 | -0.2514 | Yes |
| 76 | Ctsh | 16244 | -0.310 | -0.2595 | Yes |
| 77 | Mst1 | 16778 | -0.344 | -0.2738 | Yes |
| 78 | Usp11 | 16789 | -0.344 | -0.2616 | Yes |
| 79 | Iscu | 17063 | -0.363 | -0.2620 | Yes |
| 80 | Casp9 | 17315 | -0.382 | -0.2606 | Yes |
| 81 | Dusp6 | 17361 | -0.387 | -0.2486 | Yes |
| 82 | Ctsk | 17528 | -0.400 | -0.2422 | Yes |
| 83 | Cpq | 17563 | -0.403 | -0.2290 | Yes |
| 84 | Mmp15 | 17631 | -0.408 | -0.2173 | Yes |
| 85 | Cfd | 17795 | -0.423 | -0.2099 | Yes |
| 86 | Hmgcs2 | 17827 | -0.425 | -0.1957 | Yes |
| 87 | Sh2b2 | 17849 | -0.428 | -0.1809 | Yes |
| 88 | Bmp1 | 17995 | -0.443 | -0.1719 | Yes |
| 89 | Cd9 | 18191 | -0.465 | -0.1646 | Yes |
| 90 | Serpinb2 | 18480 | -0.503 | -0.1606 | Yes |
| 91 | Furin | 18489 | -0.506 | -0.1422 | Yes |
| 92 | Pdgfb | 18754 | -0.549 | -0.1353 | Yes |
| 93 | Gng12 | 18869 | -0.572 | -0.1199 | Yes |
| 94 | Cfb | 18978 | -0.593 | -0.1034 | Yes |
| 95 | Apoc1 | 19001 | -0.597 | -0.0824 | Yes |
| 96 | Ctse | 19243 | -0.659 | -0.0702 | Yes |
| 97 | Pf4 | 19394 | -0.716 | -0.0513 | Yes |
| 98 | Hnf4a | 19576 | -0.827 | -0.0299 | Yes |
| 99 | Rapgef3 | 19735 | -1.072 | 0.0018 | Yes |