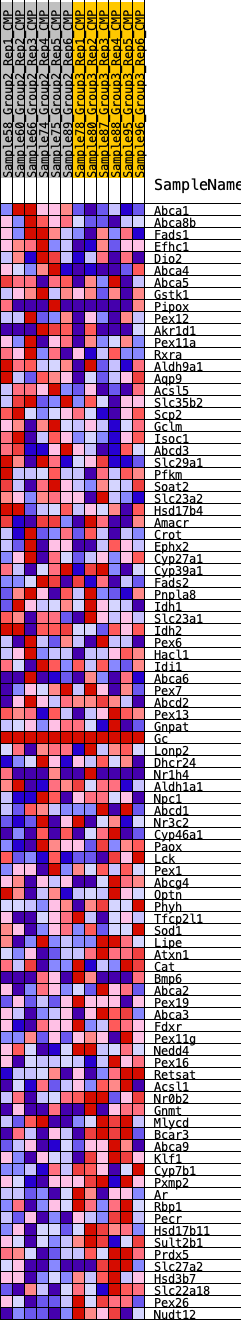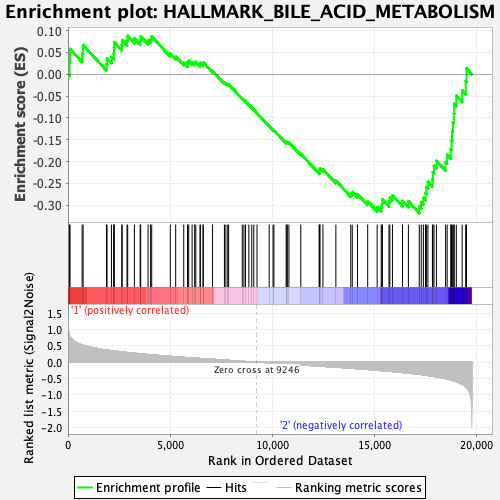
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group2_versus_Group3.CMP_Pheno.cls#Group2_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
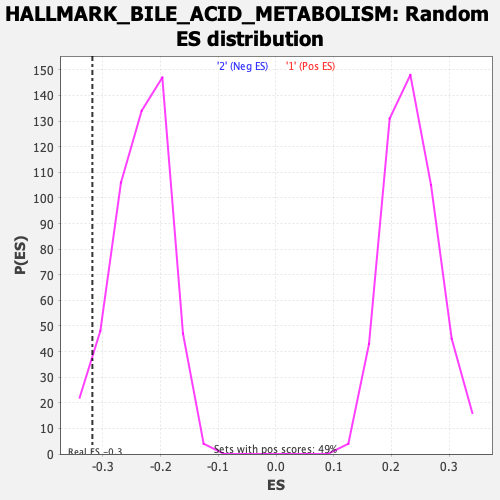
| Enrichment Score (ES) | -0.31827462 |
| Normalized Enrichment Score (NES) | -1.3564204 |
| Nominal p-value | 0.047244094 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.651 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca1 | 80 | 0.804 | 0.0278 | No |
| 2 | Abca8b | 96 | 0.782 | 0.0581 | No |
| 3 | Fads1 | 692 | 0.528 | 0.0487 | No |
| 4 | Efhc1 | 739 | 0.518 | 0.0670 | No |
| 5 | Dio2 | 1887 | 0.368 | 0.0233 | No |
| 6 | Abca4 | 1911 | 0.366 | 0.0366 | No |
| 7 | Abca5 | 2126 | 0.349 | 0.0396 | No |
| 8 | Gstk1 | 2239 | 0.341 | 0.0474 | No |
| 9 | Pipox | 2252 | 0.340 | 0.0603 | No |
| 10 | Pex12 | 2259 | 0.340 | 0.0734 | No |
| 11 | Akr1d1 | 2633 | 0.314 | 0.0669 | No |
| 12 | Pex11a | 2651 | 0.312 | 0.0784 | No |
| 13 | Rxra | 2890 | 0.296 | 0.0780 | No |
| 14 | Aldh9a1 | 2923 | 0.293 | 0.0880 | No |
| 15 | Aqp9 | 3251 | 0.273 | 0.0823 | No |
| 16 | Acsl5 | 3533 | 0.256 | 0.0782 | No |
| 17 | Slc35b2 | 3559 | 0.255 | 0.0870 | No |
| 18 | Scp2 | 3920 | 0.232 | 0.0779 | No |
| 19 | Gclm | 4043 | 0.226 | 0.0807 | No |
| 20 | Isoc1 | 4099 | 0.223 | 0.0867 | No |
| 21 | Abcd3 | 5009 | 0.178 | 0.0475 | No |
| 22 | Slc29a1 | 5272 | 0.166 | 0.0408 | No |
| 23 | Pfkm | 5667 | 0.146 | 0.0266 | No |
| 24 | Soat2 | 5855 | 0.140 | 0.0226 | No |
| 25 | Slc23a2 | 5859 | 0.139 | 0.0280 | No |
| 26 | Hsd17b4 | 5910 | 0.137 | 0.0309 | No |
| 27 | Amacr | 6076 | 0.130 | 0.0277 | No |
| 28 | Crot | 6201 | 0.124 | 0.0263 | No |
| 29 | Ephx2 | 6258 | 0.122 | 0.0283 | No |
| 30 | Cyp27a1 | 6468 | 0.113 | 0.0221 | No |
| 31 | Cyp39a1 | 6476 | 0.112 | 0.0262 | No |
| 32 | Fads2 | 6612 | 0.107 | 0.0236 | No |
| 33 | Pnpla8 | 6623 | 0.106 | 0.0273 | No |
| 34 | Idh1 | 7078 | 0.087 | 0.0077 | No |
| 35 | Slc23a1 | 7659 | 0.062 | -0.0193 | No |
| 36 | Idh2 | 7730 | 0.060 | -0.0205 | No |
| 37 | Pex6 | 7823 | 0.056 | -0.0230 | No |
| 38 | Hacl1 | 7864 | 0.055 | -0.0228 | No |
| 39 | Idi1 | 8533 | 0.026 | -0.0557 | No |
| 40 | Abca6 | 8613 | 0.023 | -0.0589 | No |
| 41 | Pex7 | 8691 | 0.020 | -0.0620 | No |
| 42 | Abcd2 | 8855 | 0.013 | -0.0698 | No |
| 43 | Pex13 | 8997 | 0.008 | -0.0766 | No |
| 44 | Gnpat | 9096 | 0.004 | -0.0814 | No |
| 45 | Gc | 9255 | 0.000 | -0.0895 | No |
| 46 | Lonp2 | 9850 | -0.020 | -0.1188 | No |
| 47 | Dhcr24 | 10038 | -0.029 | -0.1272 | No |
| 48 | Nr1h4 | 10083 | -0.031 | -0.1282 | No |
| 49 | Aldh1a1 | 10681 | -0.054 | -0.1564 | No |
| 50 | Npc1 | 10717 | -0.056 | -0.1560 | No |
| 51 | Abcd1 | 10735 | -0.056 | -0.1546 | No |
| 52 | Nr3c2 | 10816 | -0.060 | -0.1563 | No |
| 53 | Cyp46a1 | 11397 | -0.085 | -0.1824 | No |
| 54 | Paox | 12301 | -0.122 | -0.2234 | No |
| 55 | Lck | 12306 | -0.122 | -0.2188 | No |
| 56 | Pex1 | 12340 | -0.124 | -0.2156 | No |
| 57 | Abcg4 | 12482 | -0.129 | -0.2176 | No |
| 58 | Optn | 13115 | -0.152 | -0.2437 | No |
| 59 | Phyh | 13844 | -0.184 | -0.2734 | No |
| 60 | Tfcp2l1 | 13918 | -0.188 | -0.2697 | No |
| 61 | Sod1 | 14172 | -0.201 | -0.2746 | No |
| 62 | Lipe | 14673 | -0.225 | -0.2910 | No |
| 63 | Atxn1 | 15136 | -0.248 | -0.3047 | Yes |
| 64 | Cat | 15334 | -0.259 | -0.3044 | Yes |
| 65 | Bmp6 | 15374 | -0.261 | -0.2960 | Yes |
| 66 | Abca2 | 15394 | -0.262 | -0.2866 | Yes |
| 67 | Pex19 | 15715 | -0.278 | -0.2918 | Yes |
| 68 | Abca3 | 15760 | -0.282 | -0.2829 | Yes |
| 69 | Fdxr | 15886 | -0.290 | -0.2778 | Yes |
| 70 | Pex11g | 16378 | -0.319 | -0.2901 | Yes |
| 71 | Nedd4 | 16666 | -0.337 | -0.2913 | Yes |
| 72 | Pex16 | 17198 | -0.373 | -0.3035 | Yes |
| 73 | Retsat | 17295 | -0.380 | -0.2933 | Yes |
| 74 | Acsl1 | 17400 | -0.389 | -0.2831 | Yes |
| 75 | Nr0b2 | 17504 | -0.398 | -0.2725 | Yes |
| 76 | Gnmt | 17545 | -0.401 | -0.2587 | Yes |
| 77 | Mlycd | 17621 | -0.407 | -0.2463 | Yes |
| 78 | Bcar3 | 17843 | -0.427 | -0.2406 | Yes |
| 79 | Abca9 | 17861 | -0.429 | -0.2245 | Yes |
| 80 | Klf1 | 17919 | -0.435 | -0.2101 | Yes |
| 81 | Cyp7b1 | 18038 | -0.446 | -0.1984 | Yes |
| 82 | Pxmp2 | 18488 | -0.506 | -0.2011 | Yes |
| 83 | Ar | 18564 | -0.516 | -0.1845 | Yes |
| 84 | Rbp1 | 18739 | -0.546 | -0.1716 | Yes |
| 85 | Pecr | 18778 | -0.554 | -0.1516 | Yes |
| 86 | Hsd17b11 | 18803 | -0.559 | -0.1306 | Yes |
| 87 | Sult2b1 | 18843 | -0.566 | -0.1102 | Yes |
| 88 | Prdx5 | 18892 | -0.577 | -0.0898 | Yes |
| 89 | Slc27a2 | 18908 | -0.579 | -0.0676 | Yes |
| 90 | Hsd3b7 | 19012 | -0.600 | -0.0490 | Yes |
| 91 | Slc22a18 | 19298 | -0.676 | -0.0367 | Yes |
| 92 | Pex26 | 19471 | -0.754 | -0.0155 | Yes |
| 93 | Nudt12 | 19511 | -0.775 | 0.0132 | Yes |