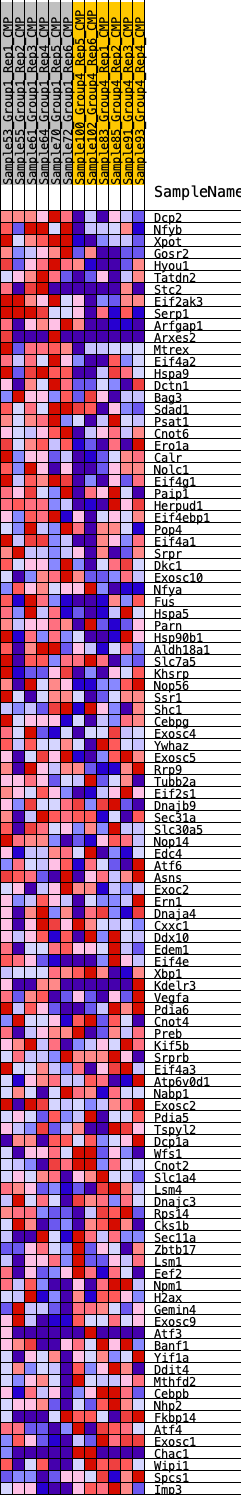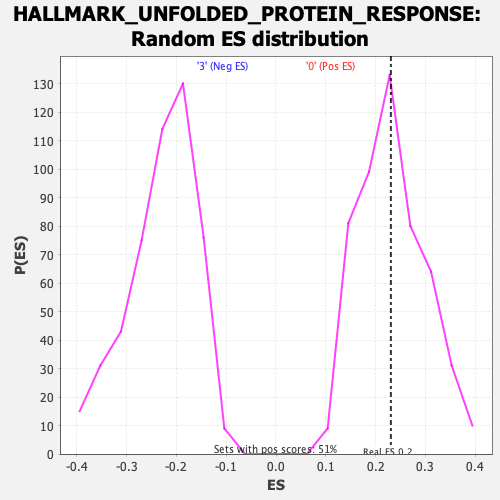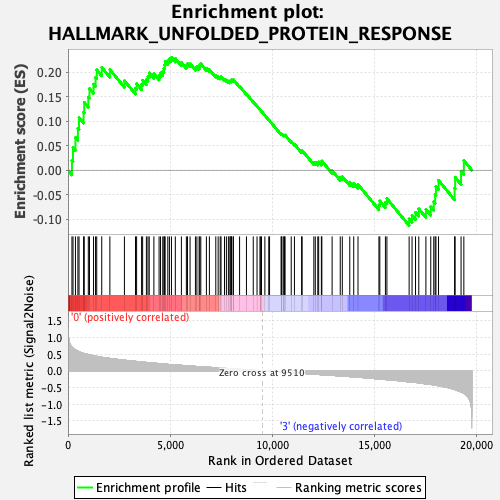
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group4.CMP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.2307382 |
| Normalized Enrichment Score (NES) | 0.9948527 |
| Nominal p-value | 0.45364892 |
| FDR q-value | 0.95225006 |
| FWER p-Value | 0.983 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dcp2 | 193 | 0.704 | 0.0201 | Yes |
| 2 | Nfyb | 245 | 0.677 | 0.0463 | Yes |
| 3 | Xpot | 364 | 0.626 | 0.0669 | Yes |
| 4 | Gosr2 | 485 | 0.591 | 0.0859 | Yes |
| 5 | Hyou1 | 540 | 0.572 | 0.1075 | Yes |
| 6 | Tatdn2 | 762 | 0.524 | 0.1185 | Yes |
| 7 | Stc2 | 795 | 0.517 | 0.1389 | Yes |
| 8 | Eif2ak3 | 994 | 0.489 | 0.1496 | Yes |
| 9 | Serp1 | 1051 | 0.479 | 0.1671 | Yes |
| 10 | Arfgap1 | 1250 | 0.451 | 0.1762 | Yes |
| 11 | Arxes2 | 1349 | 0.439 | 0.1899 | Yes |
| 12 | Mtrex | 1407 | 0.433 | 0.2054 | Yes |
| 13 | Eif4a2 | 1652 | 0.407 | 0.2103 | Yes |
| 14 | Hspa9 | 2050 | 0.370 | 0.2058 | Yes |
| 15 | Dctn1 | 2762 | 0.318 | 0.1832 | Yes |
| 16 | Bag3 | 3305 | 0.284 | 0.1677 | Yes |
| 17 | Sdad1 | 3359 | 0.280 | 0.1769 | Yes |
| 18 | Psat1 | 3603 | 0.262 | 0.1757 | Yes |
| 19 | Cnot6 | 3654 | 0.259 | 0.1842 | Yes |
| 20 | Ero1a | 3838 | 0.250 | 0.1855 | Yes |
| 21 | Calr | 3915 | 0.244 | 0.1920 | Yes |
| 22 | Nolc1 | 3976 | 0.241 | 0.1992 | Yes |
| 23 | Eif4g1 | 4205 | 0.229 | 0.1973 | Yes |
| 24 | Paip1 | 4459 | 0.216 | 0.1936 | Yes |
| 25 | Herpud1 | 4530 | 0.212 | 0.1991 | Yes |
| 26 | Eif4ebp1 | 4640 | 0.207 | 0.2024 | Yes |
| 27 | Pop4 | 4694 | 0.204 | 0.2083 | Yes |
| 28 | Eif4a1 | 4729 | 0.202 | 0.2152 | Yes |
| 29 | Srpr | 4754 | 0.200 | 0.2224 | Yes |
| 30 | Dkc1 | 4887 | 0.193 | 0.2239 | Yes |
| 31 | Exosc10 | 4965 | 0.189 | 0.2280 | Yes |
| 32 | Nfya | 5065 | 0.182 | 0.2307 | Yes |
| 33 | Fus | 5254 | 0.175 | 0.2286 | No |
| 34 | Hspa5 | 5552 | 0.163 | 0.2205 | No |
| 35 | Parn | 5795 | 0.151 | 0.2146 | No |
| 36 | Hsp90b1 | 5850 | 0.148 | 0.2181 | No |
| 37 | Aldh18a1 | 5976 | 0.143 | 0.2178 | No |
| 38 | Slc7a5 | 6239 | 0.132 | 0.2101 | No |
| 39 | Khsrp | 6311 | 0.128 | 0.2119 | No |
| 40 | Nop56 | 6420 | 0.122 | 0.2116 | No |
| 41 | Ssr1 | 6452 | 0.121 | 0.2152 | No |
| 42 | Shc1 | 6502 | 0.119 | 0.2177 | No |
| 43 | Cebpg | 6776 | 0.112 | 0.2086 | No |
| 44 | Exosc4 | 6923 | 0.105 | 0.2057 | No |
| 45 | Ywhaz | 7236 | 0.092 | 0.1937 | No |
| 46 | Exosc5 | 7352 | 0.087 | 0.1916 | No |
| 47 | Rrp9 | 7451 | 0.083 | 0.1901 | No |
| 48 | Tubb2a | 7492 | 0.082 | 0.1915 | No |
| 49 | Eif2s1 | 7662 | 0.075 | 0.1861 | No |
| 50 | Dnajb9 | 7755 | 0.070 | 0.1844 | No |
| 51 | Sec31a | 7853 | 0.066 | 0.1823 | No |
| 52 | Slc30a5 | 7932 | 0.062 | 0.1810 | No |
| 53 | Nop14 | 7942 | 0.062 | 0.1832 | No |
| 54 | Edc4 | 7996 | 0.060 | 0.1830 | No |
| 55 | Atf6 | 8005 | 0.059 | 0.1851 | No |
| 56 | Asns | 8088 | 0.056 | 0.1833 | No |
| 57 | Exoc2 | 8094 | 0.056 | 0.1855 | No |
| 58 | Ern1 | 8401 | 0.044 | 0.1718 | No |
| 59 | Dnaja4 | 8737 | 0.030 | 0.1560 | No |
| 60 | Cxxc1 | 9070 | 0.018 | 0.1399 | No |
| 61 | Ddx10 | 9255 | 0.010 | 0.1310 | No |
| 62 | Edem1 | 9408 | 0.004 | 0.1234 | No |
| 63 | Eif4e | 9411 | 0.004 | 0.1235 | No |
| 64 | Xbp1 | 9474 | 0.002 | 0.1204 | No |
| 65 | Kdelr3 | 9639 | -0.000 | 0.1121 | No |
| 66 | Vegfa | 9830 | -0.009 | 0.1028 | No |
| 67 | Pdia6 | 9868 | -0.010 | 0.1013 | No |
| 68 | Cnot4 | 10438 | -0.031 | 0.0737 | No |
| 69 | Preb | 10445 | -0.032 | 0.0747 | No |
| 70 | Kif5b | 10529 | -0.035 | 0.0720 | No |
| 71 | Srprb | 10571 | -0.037 | 0.0715 | No |
| 72 | Eif4a3 | 10599 | -0.038 | 0.0717 | No |
| 73 | Atp6v0d1 | 10628 | -0.039 | 0.0719 | No |
| 74 | Nabp1 | 10929 | -0.051 | 0.0588 | No |
| 75 | Exosc2 | 11086 | -0.058 | 0.0533 | No |
| 76 | Pdia5 | 11437 | -0.072 | 0.0386 | No |
| 77 | Tspyl2 | 11463 | -0.073 | 0.0405 | No |
| 78 | Dcp1a | 12037 | -0.098 | 0.0155 | No |
| 79 | Wfs1 | 12107 | -0.100 | 0.0162 | No |
| 80 | Cnot2 | 12230 | -0.107 | 0.0146 | No |
| 81 | Slc1a4 | 12273 | -0.108 | 0.0170 | No |
| 82 | Lsm4 | 12405 | -0.114 | 0.0152 | No |
| 83 | Dnajc3 | 12429 | -0.116 | 0.0190 | No |
| 84 | Rps14 | 12931 | -0.135 | -0.0008 | No |
| 85 | Cks1b | 13329 | -0.151 | -0.0146 | No |
| 86 | Sec11a | 13430 | -0.156 | -0.0130 | No |
| 87 | Zbtb17 | 13796 | -0.172 | -0.0243 | No |
| 88 | Lsm1 | 13989 | -0.177 | -0.0265 | No |
| 89 | Eef2 | 14199 | -0.188 | -0.0291 | No |
| 90 | Npm1 | 15224 | -0.242 | -0.0709 | No |
| 91 | H2ax | 15267 | -0.243 | -0.0628 | No |
| 92 | Gemin4 | 15541 | -0.260 | -0.0656 | No |
| 93 | Exosc9 | 15614 | -0.264 | -0.0580 | No |
| 94 | Atf3 | 16702 | -0.329 | -0.0993 | No |
| 95 | Banf1 | 16845 | -0.335 | -0.0923 | No |
| 96 | Yif1a | 17015 | -0.346 | -0.0862 | No |
| 97 | Ddit4 | 17175 | -0.360 | -0.0790 | No |
| 98 | Mthfd2 | 17523 | -0.389 | -0.0801 | No |
| 99 | Cebpb | 17761 | -0.407 | -0.0748 | No |
| 100 | Nhp2 | 17908 | -0.420 | -0.0644 | No |
| 101 | Fkbp14 | 17975 | -0.427 | -0.0496 | No |
| 102 | Atf4 | 18015 | -0.432 | -0.0332 | No |
| 103 | Exosc1 | 18141 | -0.444 | -0.0207 | No |
| 104 | Chac1 | 18928 | -0.559 | -0.0369 | No |
| 105 | Wipi1 | 18954 | -0.563 | -0.0142 | No |
| 106 | Spcs1 | 19245 | -0.635 | -0.0020 | No |
| 107 | Imp3 | 19381 | -0.675 | 0.0198 | No |