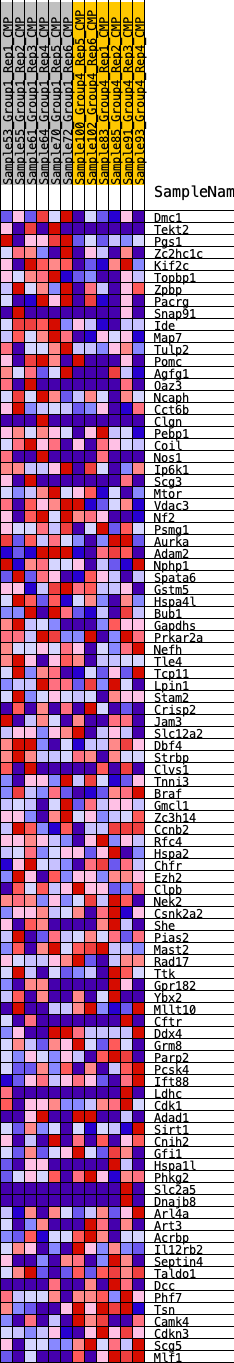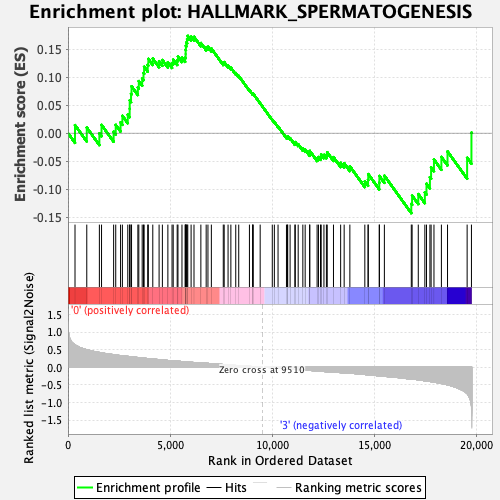
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group4.CMP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.17375825 |
| Normalized Enrichment Score (NES) | 0.76743406 |
| Nominal p-value | 0.8389662 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dmc1 | 343 | 0.635 | 0.0143 | Yes |
| 2 | Tekt2 | 919 | 0.497 | 0.0100 | Yes |
| 3 | Pgs1 | 1536 | 0.419 | -0.0004 | Yes |
| 4 | Zc2hc1c | 1642 | 0.408 | 0.0147 | Yes |
| 5 | Kif2c | 2234 | 0.357 | 0.0025 | Yes |
| 6 | Topbp1 | 2337 | 0.349 | 0.0148 | Yes |
| 7 | Zpbp | 2573 | 0.330 | 0.0193 | Yes |
| 8 | Pacrg | 2663 | 0.323 | 0.0310 | Yes |
| 9 | Snap91 | 2927 | 0.308 | 0.0330 | Yes |
| 10 | Ide | 3019 | 0.302 | 0.0435 | Yes |
| 11 | Map7 | 3022 | 0.302 | 0.0585 | Yes |
| 12 | Tulp2 | 3087 | 0.297 | 0.0701 | Yes |
| 13 | Pomc | 3111 | 0.295 | 0.0837 | Yes |
| 14 | Agfg1 | 3421 | 0.275 | 0.0817 | Yes |
| 15 | Oaz3 | 3467 | 0.271 | 0.0930 | Yes |
| 16 | Ncaph | 3632 | 0.261 | 0.0977 | Yes |
| 17 | Cct6b | 3697 | 0.257 | 0.1073 | Yes |
| 18 | Clgn | 3727 | 0.255 | 0.1186 | Yes |
| 19 | Pebp1 | 3903 | 0.245 | 0.1220 | Yes |
| 20 | Coil | 3933 | 0.243 | 0.1327 | Yes |
| 21 | Nos1 | 4151 | 0.232 | 0.1333 | Yes |
| 22 | Ip6k1 | 4460 | 0.216 | 0.1284 | Yes |
| 23 | Scg3 | 4621 | 0.207 | 0.1307 | Yes |
| 24 | Mtor | 4890 | 0.192 | 0.1267 | Yes |
| 25 | Vdac3 | 5094 | 0.181 | 0.1254 | Yes |
| 26 | Nf2 | 5151 | 0.179 | 0.1315 | Yes |
| 27 | Psmg1 | 5346 | 0.171 | 0.1302 | Yes |
| 28 | Aurka | 5384 | 0.170 | 0.1368 | Yes |
| 29 | Adam2 | 5580 | 0.162 | 0.1350 | Yes |
| 30 | Nphp1 | 5733 | 0.153 | 0.1349 | Yes |
| 31 | Spata6 | 5760 | 0.152 | 0.1412 | Yes |
| 32 | Gstm5 | 5761 | 0.152 | 0.1489 | Yes |
| 33 | Hspa4l | 5772 | 0.152 | 0.1559 | Yes |
| 34 | Bub1 | 5798 | 0.151 | 0.1622 | Yes |
| 35 | Gapdhs | 5833 | 0.149 | 0.1679 | Yes |
| 36 | Prkar2a | 5864 | 0.147 | 0.1738 | Yes |
| 37 | Nefh | 6027 | 0.141 | 0.1726 | No |
| 38 | Tle4 | 6169 | 0.134 | 0.1721 | No |
| 39 | Tcp11 | 6505 | 0.118 | 0.1610 | No |
| 40 | Lpin1 | 6765 | 0.112 | 0.1534 | No |
| 41 | Stam2 | 6852 | 0.109 | 0.1545 | No |
| 42 | Crisp2 | 7019 | 0.101 | 0.1512 | No |
| 43 | Jam3 | 7602 | 0.077 | 0.1254 | No |
| 44 | Slc12a2 | 7647 | 0.076 | 0.1270 | No |
| 45 | Dbf4 | 7837 | 0.067 | 0.1207 | No |
| 46 | Strbp | 7974 | 0.061 | 0.1168 | No |
| 47 | Clvs1 | 8213 | 0.051 | 0.1073 | No |
| 48 | Tnni3 | 8359 | 0.046 | 0.1022 | No |
| 49 | Braf | 8883 | 0.025 | 0.0769 | No |
| 50 | Gmcl1 | 9041 | 0.019 | 0.0698 | No |
| 51 | Zc3h14 | 9047 | 0.019 | 0.0705 | No |
| 52 | Ccnb2 | 9065 | 0.018 | 0.0705 | No |
| 53 | Rfc4 | 9409 | 0.004 | 0.0533 | No |
| 54 | Hspa2 | 10002 | -0.015 | 0.0240 | No |
| 55 | Chfr | 10103 | -0.018 | 0.0198 | No |
| 56 | Ezh2 | 10284 | -0.025 | 0.0119 | No |
| 57 | Clpb | 10696 | -0.041 | -0.0069 | No |
| 58 | Nek2 | 10743 | -0.043 | -0.0071 | No |
| 59 | Csnk2a2 | 10757 | -0.044 | -0.0056 | No |
| 60 | She | 10874 | -0.049 | -0.0090 | No |
| 61 | Pias2 | 11108 | -0.058 | -0.0179 | No |
| 62 | Mast2 | 11128 | -0.059 | -0.0159 | No |
| 63 | Rad17 | 11270 | -0.065 | -0.0198 | No |
| 64 | Ttk | 11498 | -0.075 | -0.0276 | No |
| 65 | Gpr182 | 11612 | -0.080 | -0.0293 | No |
| 66 | Ybx2 | 11833 | -0.089 | -0.0360 | No |
| 67 | Mllt10 | 11836 | -0.090 | -0.0317 | No |
| 68 | Cftr | 12196 | -0.105 | -0.0447 | No |
| 69 | Ddx4 | 12263 | -0.108 | -0.0426 | No |
| 70 | Grm8 | 12363 | -0.112 | -0.0420 | No |
| 71 | Parp2 | 12396 | -0.114 | -0.0380 | No |
| 72 | Pcsk4 | 12531 | -0.121 | -0.0387 | No |
| 73 | Ift88 | 12652 | -0.125 | -0.0386 | No |
| 74 | Ldhc | 12692 | -0.127 | -0.0342 | No |
| 75 | Cdk1 | 12998 | -0.137 | -0.0428 | No |
| 76 | Adad1 | 13349 | -0.152 | -0.0530 | No |
| 77 | Sirt1 | 13521 | -0.160 | -0.0537 | No |
| 78 | Cnih2 | 13800 | -0.172 | -0.0592 | No |
| 79 | Gfi1 | 14533 | -0.206 | -0.0861 | No |
| 80 | Hspa1l | 14691 | -0.214 | -0.0834 | No |
| 81 | Phkg2 | 14701 | -0.215 | -0.0731 | No |
| 82 | Slc2a5 | 15240 | -0.242 | -0.0883 | No |
| 83 | Dnajb8 | 15246 | -0.242 | -0.0765 | No |
| 84 | Arl4a | 15490 | -0.257 | -0.0759 | No |
| 85 | Art3 | 16808 | -0.333 | -0.1262 | No |
| 86 | Acrbp | 16848 | -0.335 | -0.1114 | No |
| 87 | Il12rb2 | 17150 | -0.358 | -0.1088 | No |
| 88 | Septin4 | 17472 | -0.386 | -0.1058 | No |
| 89 | Taldo1 | 17553 | -0.391 | -0.0903 | No |
| 90 | Dcc | 17720 | -0.403 | -0.0786 | No |
| 91 | Phf7 | 17778 | -0.409 | -0.0610 | No |
| 92 | Tsn | 17918 | -0.421 | -0.0470 | No |
| 93 | Camk4 | 18282 | -0.460 | -0.0425 | No |
| 94 | Cdkn3 | 18582 | -0.499 | -0.0327 | No |
| 95 | Scg5 | 19542 | -0.749 | -0.0440 | No |
| 96 | Mlf1 | 19753 | -1.110 | 0.0009 | No |