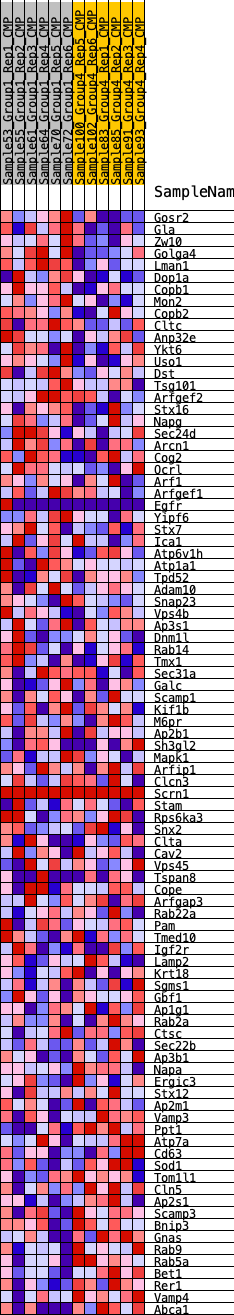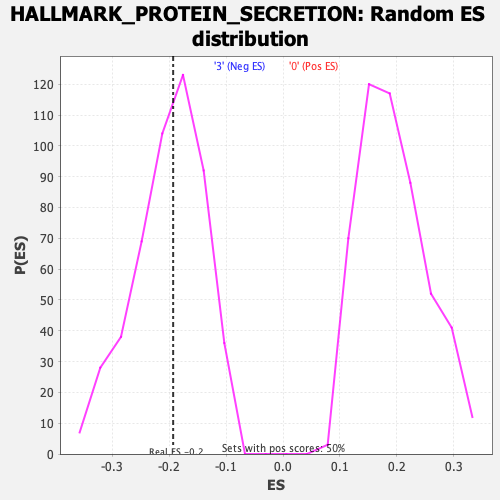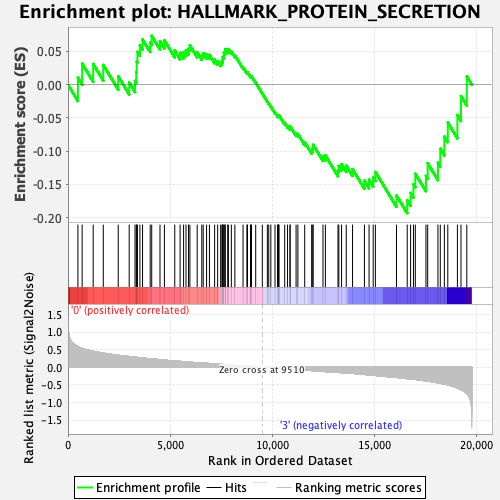
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group4.CMP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.19328777 |
| Normalized Enrichment Score (NES) | -0.9698409 |
| Nominal p-value | 0.49496982 |
| FDR q-value | 0.6403512 |
| FWER p-Value | 0.984 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gosr2 | 485 | 0.591 | 0.0104 | No |
| 2 | Gla | 693 | 0.537 | 0.0317 | No |
| 3 | Zw10 | 1233 | 0.453 | 0.0311 | No |
| 4 | Golga4 | 1727 | 0.399 | 0.0297 | No |
| 5 | Lman1 | 2459 | 0.338 | 0.0126 | No |
| 6 | Dop1a | 2996 | 0.304 | 0.0034 | No |
| 7 | Copb1 | 3282 | 0.285 | 0.0058 | No |
| 8 | Mon2 | 3346 | 0.280 | 0.0192 | No |
| 9 | Copb2 | 3364 | 0.279 | 0.0349 | No |
| 10 | Cltc | 3400 | 0.276 | 0.0495 | No |
| 11 | Anp32e | 3521 | 0.266 | 0.0592 | No |
| 12 | Ykt6 | 3650 | 0.260 | 0.0681 | No |
| 13 | Uso1 | 4024 | 0.239 | 0.0633 | No |
| 14 | Dst | 4099 | 0.235 | 0.0734 | No |
| 15 | Tsg101 | 4504 | 0.214 | 0.0656 | No |
| 16 | Arfgef2 | 4720 | 0.202 | 0.0666 | No |
| 17 | Stx16 | 5227 | 0.177 | 0.0514 | No |
| 18 | Napg | 5488 | 0.166 | 0.0480 | No |
| 19 | Sec24d | 5659 | 0.158 | 0.0488 | No |
| 20 | Arcn1 | 5777 | 0.152 | 0.0518 | No |
| 21 | Cog2 | 5902 | 0.145 | 0.0541 | No |
| 22 | Ocrl | 5968 | 0.143 | 0.0593 | No |
| 23 | Arf1 | 6325 | 0.127 | 0.0487 | No |
| 24 | Arfgef1 | 6550 | 0.117 | 0.0443 | No |
| 25 | Egfr | 6620 | 0.114 | 0.0475 | No |
| 26 | Yipf6 | 6785 | 0.112 | 0.0458 | No |
| 27 | Stx7 | 6924 | 0.105 | 0.0450 | No |
| 28 | Ica1 | 7179 | 0.094 | 0.0377 | No |
| 29 | Atp6v1h | 7327 | 0.089 | 0.0355 | No |
| 30 | Atp1a1 | 7474 | 0.082 | 0.0329 | No |
| 31 | Tpd52 | 7535 | 0.080 | 0.0346 | No |
| 32 | Adam10 | 7567 | 0.078 | 0.0377 | No |
| 33 | Snap23 | 7579 | 0.078 | 0.0417 | No |
| 34 | Vps4b | 7633 | 0.076 | 0.0435 | No |
| 35 | Ap3s1 | 7634 | 0.076 | 0.0481 | No |
| 36 | Dnm1l | 7689 | 0.073 | 0.0496 | No |
| 37 | Rab14 | 7697 | 0.073 | 0.0536 | No |
| 38 | Tmx1 | 7821 | 0.067 | 0.0514 | No |
| 39 | Sec31a | 7853 | 0.066 | 0.0537 | No |
| 40 | Galc | 7997 | 0.060 | 0.0499 | No |
| 41 | Scamp1 | 8170 | 0.052 | 0.0443 | No |
| 42 | Kif1b | 8570 | 0.037 | 0.0262 | No |
| 43 | M6pr | 8759 | 0.029 | 0.0184 | No |
| 44 | Ap2b1 | 8788 | 0.028 | 0.0187 | No |
| 45 | Sh3gl2 | 8934 | 0.023 | 0.0127 | No |
| 46 | Mapk1 | 8957 | 0.022 | 0.0129 | No |
| 47 | Arfip1 | 8986 | 0.021 | 0.0127 | No |
| 48 | Clcn3 | 9190 | 0.012 | 0.0031 | No |
| 49 | Scrn1 | 9514 | 0.000 | -0.0133 | No |
| 50 | Stam | 9760 | -0.005 | -0.0254 | No |
| 51 | Rps6ka3 | 9828 | -0.009 | -0.0283 | No |
| 52 | Snx2 | 9931 | -0.012 | -0.0328 | No |
| 53 | Clta | 10135 | -0.019 | -0.0420 | No |
| 54 | Cav2 | 10259 | -0.024 | -0.0468 | No |
| 55 | Vps45 | 10300 | -0.025 | -0.0473 | No |
| 56 | Tspan8 | 10302 | -0.025 | -0.0459 | No |
| 57 | Cope | 10334 | -0.027 | -0.0458 | No |
| 58 | Arfgap3 | 10610 | -0.038 | -0.0576 | No |
| 59 | Rab22a | 10749 | -0.044 | -0.0620 | No |
| 60 | Pam | 10870 | -0.049 | -0.0652 | No |
| 61 | Tmed10 | 10873 | -0.049 | -0.0624 | No |
| 62 | Igf2r | 11167 | -0.061 | -0.0736 | No |
| 63 | Lamp2 | 11255 | -0.065 | -0.0742 | No |
| 64 | Krt18 | 11589 | -0.079 | -0.0864 | No |
| 65 | Sgms1 | 11929 | -0.094 | -0.0981 | No |
| 66 | Gbf1 | 11985 | -0.096 | -0.0952 | No |
| 67 | Ap1g1 | 12001 | -0.096 | -0.0903 | No |
| 68 | Rab2a | 12484 | -0.118 | -0.1078 | No |
| 69 | Ctsc | 12602 | -0.123 | -0.1064 | No |
| 70 | Sec22b | 13219 | -0.147 | -0.1290 | No |
| 71 | Ap3b1 | 13260 | -0.149 | -0.1222 | No |
| 72 | Napa | 13394 | -0.154 | -0.1198 | No |
| 73 | Ergic3 | 13623 | -0.165 | -0.1217 | No |
| 74 | Stx12 | 13932 | -0.174 | -0.1270 | No |
| 75 | Ap2m1 | 14513 | -0.205 | -0.1443 | No |
| 76 | Vamp3 | 14740 | -0.217 | -0.1430 | No |
| 77 | Ppt1 | 14942 | -0.227 | -0.1397 | No |
| 78 | Atp7a | 15049 | -0.234 | -0.1313 | No |
| 79 | Cd63 | 16083 | -0.288 | -0.1667 | No |
| 80 | Sod1 | 16608 | -0.323 | -0.1741 | Yes |
| 81 | Tom1l1 | 16774 | -0.332 | -0.1629 | Yes |
| 82 | Cln5 | 16915 | -0.340 | -0.1498 | Yes |
| 83 | Ap2s1 | 17005 | -0.345 | -0.1339 | Yes |
| 84 | Scamp3 | 17526 | -0.389 | -0.1373 | Yes |
| 85 | Bnip3 | 17611 | -0.394 | -0.1182 | Yes |
| 86 | Gnas | 18113 | -0.442 | -0.1174 | Yes |
| 87 | Rab9 | 18228 | -0.453 | -0.0964 | Yes |
| 88 | Rab5a | 18427 | -0.478 | -0.0781 | Yes |
| 89 | Bet1 | 18595 | -0.500 | -0.0569 | Yes |
| 90 | Rer1 | 19066 | -0.590 | -0.0458 | Yes |
| 91 | Vamp4 | 19241 | -0.634 | -0.0171 | Yes |
| 92 | Abca1 | 19526 | -0.742 | 0.0124 | Yes |