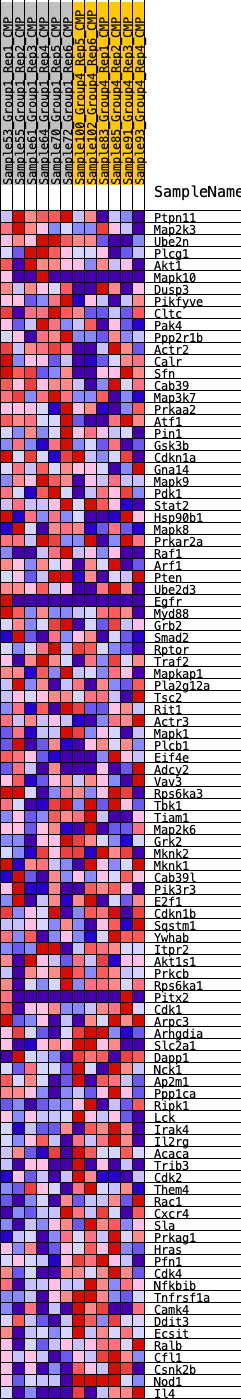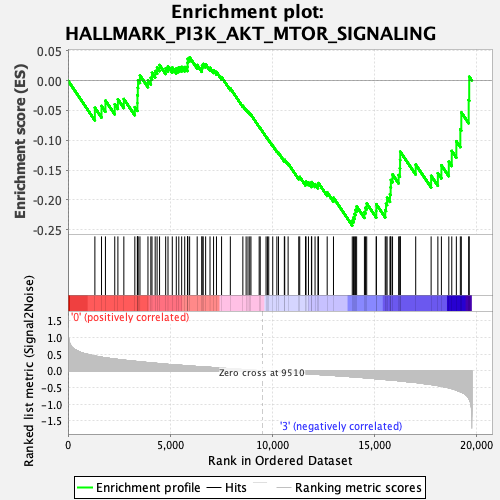
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group4.CMP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.24308288 |
| Normalized Enrichment Score (NES) | -1.1712183 |
| Nominal p-value | 0.26003823 |
| FDR q-value | 0.4113022 |
| FWER p-Value | 0.893 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ptpn11 | 1316 | 0.444 | -0.0456 | No |
| 2 | Map2k3 | 1641 | 0.408 | -0.0425 | No |
| 3 | Ube2n | 1833 | 0.389 | -0.0336 | No |
| 4 | Plcg1 | 2289 | 0.352 | -0.0399 | No |
| 5 | Akt1 | 2443 | 0.339 | -0.0314 | No |
| 6 | Mapk10 | 2734 | 0.318 | -0.0309 | No |
| 7 | Dusp3 | 3272 | 0.286 | -0.0445 | No |
| 8 | Pikfyve | 3398 | 0.277 | -0.0376 | No |
| 9 | Cltc | 3400 | 0.276 | -0.0244 | No |
| 10 | Pak4 | 3418 | 0.275 | -0.0121 | No |
| 11 | Ppp2r1b | 3430 | 0.274 | 0.0004 | No |
| 12 | Actr2 | 3523 | 0.266 | 0.0085 | No |
| 13 | Calr | 3915 | 0.244 | 0.0004 | No |
| 14 | Sfn | 4056 | 0.237 | 0.0046 | No |
| 15 | Cab39 | 4112 | 0.234 | 0.0130 | No |
| 16 | Map3k7 | 4268 | 0.226 | 0.0160 | No |
| 17 | Prkaa2 | 4362 | 0.221 | 0.0218 | No |
| 18 | Atf1 | 4483 | 0.215 | 0.0260 | No |
| 19 | Pin1 | 4782 | 0.199 | 0.0204 | No |
| 20 | Gsk3b | 4896 | 0.192 | 0.0239 | No |
| 21 | Cdkn1a | 5106 | 0.180 | 0.0219 | No |
| 22 | Gna14 | 5302 | 0.173 | 0.0203 | No |
| 23 | Mapk9 | 5425 | 0.168 | 0.0221 | No |
| 24 | Pdk1 | 5565 | 0.162 | 0.0228 | No |
| 25 | Stat2 | 5715 | 0.154 | 0.0226 | No |
| 26 | Hsp90b1 | 5850 | 0.148 | 0.0229 | No |
| 27 | Mapk8 | 5856 | 0.148 | 0.0297 | No |
| 28 | Prkar2a | 5864 | 0.147 | 0.0365 | No |
| 29 | Raf1 | 5955 | 0.143 | 0.0387 | No |
| 30 | Arf1 | 6325 | 0.127 | 0.0261 | No |
| 31 | Pten | 6552 | 0.117 | 0.0202 | No |
| 32 | Ube2d3 | 6565 | 0.116 | 0.0251 | No |
| 33 | Egfr | 6620 | 0.114 | 0.0278 | No |
| 34 | Myd88 | 6739 | 0.113 | 0.0273 | No |
| 35 | Grb2 | 6953 | 0.104 | 0.0214 | No |
| 36 | Smad2 | 7127 | 0.097 | 0.0173 | No |
| 37 | Rptor | 7269 | 0.091 | 0.0144 | No |
| 38 | Traf2 | 7519 | 0.081 | 0.0056 | No |
| 39 | Mapkap1 | 7948 | 0.062 | -0.0131 | No |
| 40 | Pla2g12a | 8561 | 0.038 | -0.0424 | No |
| 41 | Tsc2 | 8721 | 0.031 | -0.0490 | No |
| 42 | Rit1 | 8813 | 0.027 | -0.0523 | No |
| 43 | Actr3 | 8899 | 0.024 | -0.0555 | No |
| 44 | Mapk1 | 8957 | 0.022 | -0.0573 | No |
| 45 | Plcb1 | 9360 | 0.006 | -0.0775 | No |
| 46 | Eif4e | 9411 | 0.004 | -0.0798 | No |
| 47 | Adcy2 | 9697 | -0.003 | -0.0942 | No |
| 48 | Vav3 | 9764 | -0.005 | -0.0973 | No |
| 49 | Rps6ka3 | 9828 | -0.009 | -0.1001 | No |
| 50 | Tbk1 | 10024 | -0.015 | -0.1092 | No |
| 51 | Tiam1 | 10226 | -0.023 | -0.1184 | No |
| 52 | Map2k6 | 10305 | -0.026 | -0.1211 | No |
| 53 | Grk2 | 10592 | -0.038 | -0.1338 | No |
| 54 | Mknk2 | 10611 | -0.038 | -0.1329 | No |
| 55 | Mknk1 | 10777 | -0.045 | -0.1391 | No |
| 56 | Cab39l | 11298 | -0.066 | -0.1624 | No |
| 57 | Pik3r3 | 11346 | -0.068 | -0.1615 | No |
| 58 | E2f1 | 11628 | -0.081 | -0.1719 | No |
| 59 | Cdkn1b | 11654 | -0.082 | -0.1692 | No |
| 60 | Sqstm1 | 11773 | -0.086 | -0.1711 | No |
| 61 | Ywhab | 11919 | -0.093 | -0.1740 | No |
| 62 | Itpr2 | 11932 | -0.094 | -0.1701 | No |
| 63 | Akt1s1 | 12096 | -0.100 | -0.1736 | No |
| 64 | Prkcb | 12237 | -0.107 | -0.1756 | No |
| 65 | Rps6ka1 | 12264 | -0.108 | -0.1718 | No |
| 66 | Pitx2 | 12690 | -0.127 | -0.1873 | No |
| 67 | Cdk1 | 12998 | -0.137 | -0.1964 | No |
| 68 | Arpc3 | 13918 | -0.173 | -0.2348 | Yes |
| 69 | Arhgdia | 13980 | -0.176 | -0.2294 | Yes |
| 70 | Slc2a1 | 14034 | -0.179 | -0.2236 | Yes |
| 71 | Dapp1 | 14073 | -0.181 | -0.2168 | Yes |
| 72 | Nck1 | 14132 | -0.184 | -0.2109 | Yes |
| 73 | Ap2m1 | 14513 | -0.205 | -0.2204 | Yes |
| 74 | Ppp1ca | 14564 | -0.208 | -0.2130 | Yes |
| 75 | Ripk1 | 14625 | -0.210 | -0.2060 | Yes |
| 76 | Lck | 15093 | -0.236 | -0.2184 | Yes |
| 77 | Irak4 | 15096 | -0.236 | -0.2072 | Yes |
| 78 | Il2rg | 15532 | -0.259 | -0.2169 | Yes |
| 79 | Acaca | 15571 | -0.261 | -0.2063 | Yes |
| 80 | Trib3 | 15622 | -0.265 | -0.1962 | Yes |
| 81 | Cdk2 | 15765 | -0.270 | -0.1905 | Yes |
| 82 | Them4 | 15797 | -0.272 | -0.1790 | Yes |
| 83 | Rac1 | 15809 | -0.273 | -0.1665 | Yes |
| 84 | Cxcr4 | 15890 | -0.277 | -0.1573 | Yes |
| 85 | Sla | 16189 | -0.295 | -0.1583 | Yes |
| 86 | Prkag1 | 16250 | -0.299 | -0.1471 | Yes |
| 87 | Hras | 16259 | -0.300 | -0.1331 | Yes |
| 88 | Pfn1 | 16266 | -0.300 | -0.1190 | Yes |
| 89 | Cdk4 | 17024 | -0.347 | -0.1409 | Yes |
| 90 | Nfkbib | 17779 | -0.409 | -0.1596 | Yes |
| 91 | Tnfrsf1a | 18109 | -0.440 | -0.1552 | Yes |
| 92 | Camk4 | 18282 | -0.460 | -0.1419 | Yes |
| 93 | Ddit3 | 18645 | -0.507 | -0.1360 | Yes |
| 94 | Ecsit | 18790 | -0.530 | -0.1179 | Yes |
| 95 | Ralb | 19014 | -0.576 | -0.1017 | Yes |
| 96 | Cfl1 | 19209 | -0.625 | -0.0816 | Yes |
| 97 | Csnk2b | 19257 | -0.638 | -0.0534 | Yes |
| 98 | Nod1 | 19618 | -0.815 | -0.0327 | Yes |
| 99 | Il4 | 19644 | -0.844 | 0.0065 | Yes |