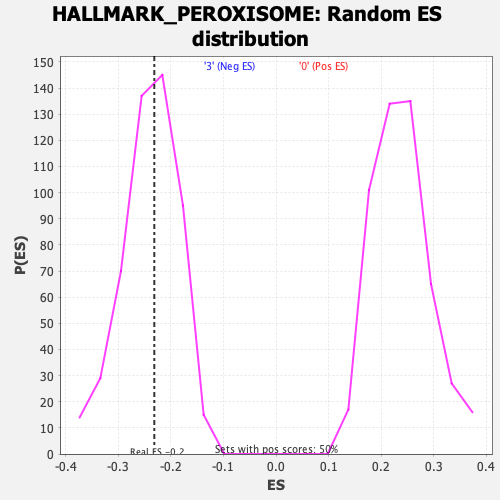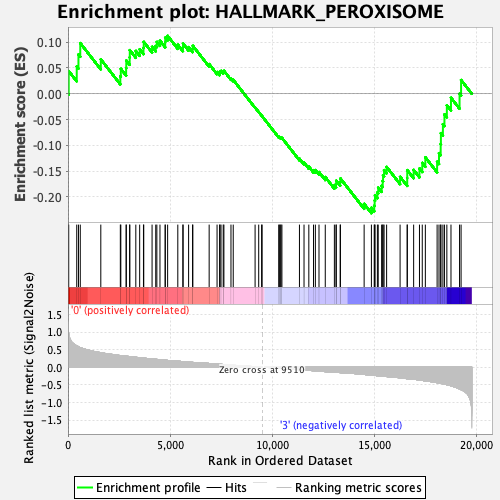
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group4.CMP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.23157968 |
| Normalized Enrichment Score (NES) | -0.9688693 |
| Nominal p-value | 0.5247525 |
| FDR q-value | 0.6088096 |
| FWER p-Value | 0.984 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex5 | 35 | 0.962 | 0.0440 | No |
| 2 | Gnpat | 428 | 0.607 | 0.0529 | No |
| 3 | Slc27a2 | 512 | 0.581 | 0.0763 | No |
| 4 | Hmgcl | 601 | 0.556 | 0.0983 | No |
| 5 | Pex11a | 1606 | 0.411 | 0.0668 | No |
| 6 | Dhcr24 | 2562 | 0.331 | 0.0340 | No |
| 7 | Ywhah | 2586 | 0.328 | 0.0484 | No |
| 8 | Acsl5 | 2846 | 0.312 | 0.0501 | No |
| 9 | Abcb9 | 2854 | 0.311 | 0.0645 | No |
| 10 | Ide | 3019 | 0.302 | 0.0705 | No |
| 11 | Aldh9a1 | 3025 | 0.302 | 0.0846 | No |
| 12 | Slc23a2 | 3320 | 0.283 | 0.0831 | No |
| 13 | Acot8 | 3509 | 0.268 | 0.0863 | No |
| 14 | Smarcc1 | 3695 | 0.257 | 0.0891 | No |
| 15 | Isoc1 | 3709 | 0.257 | 0.1007 | No |
| 16 | Pex6 | 4116 | 0.234 | 0.0912 | No |
| 17 | Abcc5 | 4296 | 0.225 | 0.0928 | No |
| 18 | Acsl4 | 4350 | 0.222 | 0.1006 | No |
| 19 | Acsl1 | 4503 | 0.214 | 0.1030 | No |
| 20 | Msh2 | 4758 | 0.200 | 0.0996 | No |
| 21 | Itgb1bp1 | 4761 | 0.200 | 0.1090 | No |
| 22 | Elovl5 | 4878 | 0.193 | 0.1123 | No |
| 23 | Top2a | 5376 | 0.170 | 0.0951 | No |
| 24 | Cln6 | 5626 | 0.160 | 0.0901 | No |
| 25 | Cln8 | 5633 | 0.159 | 0.0973 | No |
| 26 | Hsd11b2 | 5905 | 0.145 | 0.0905 | No |
| 27 | Idh1 | 6098 | 0.137 | 0.0872 | No |
| 28 | Lonp2 | 6112 | 0.136 | 0.0931 | No |
| 29 | Idh2 | 6911 | 0.106 | 0.0576 | No |
| 30 | Acox1 | 7298 | 0.089 | 0.0422 | No |
| 31 | Idi1 | 7423 | 0.084 | 0.0399 | No |
| 32 | Crat | 7440 | 0.083 | 0.0431 | No |
| 33 | Cnbp | 7491 | 0.082 | 0.0444 | No |
| 34 | Ercc3 | 7599 | 0.077 | 0.0426 | No |
| 35 | Vps4b | 7633 | 0.076 | 0.0446 | No |
| 36 | Siah1a | 7979 | 0.061 | 0.0299 | No |
| 37 | Fads1 | 8090 | 0.056 | 0.0270 | No |
| 38 | Abcd1 | 9160 | 0.013 | -0.0266 | No |
| 39 | Aldh1a1 | 9337 | 0.007 | -0.0352 | No |
| 40 | Pabpc1 | 9479 | 0.001 | -0.0423 | No |
| 41 | Sema3c | 9504 | 0.000 | -0.0435 | No |
| 42 | Mlycd | 10318 | -0.026 | -0.0836 | No |
| 43 | Ephx2 | 10348 | -0.027 | -0.0838 | No |
| 44 | Rdh11 | 10394 | -0.029 | -0.0847 | No |
| 45 | Abcd2 | 10429 | -0.031 | -0.0849 | No |
| 46 | Cacna1b | 10473 | -0.033 | -0.0856 | No |
| 47 | Abcb4 | 11332 | -0.068 | -0.1259 | No |
| 48 | Ercc1 | 11556 | -0.078 | -0.1335 | No |
| 49 | Hsd17b4 | 11791 | -0.087 | -0.1413 | No |
| 50 | Fdps | 12021 | -0.097 | -0.1483 | No |
| 51 | Cat | 12111 | -0.101 | -0.1480 | No |
| 52 | Ctbp1 | 12289 | -0.109 | -0.1518 | No |
| 53 | Abcd3 | 12596 | -0.123 | -0.1615 | No |
| 54 | Ehhadh | 13042 | -0.140 | -0.1775 | No |
| 55 | Ech1 | 13124 | -0.143 | -0.1748 | No |
| 56 | Dhrs3 | 13134 | -0.143 | -0.1685 | No |
| 57 | Slc25a19 | 13327 | -0.151 | -0.1710 | No |
| 58 | Pex14 | 13338 | -0.152 | -0.1643 | No |
| 59 | Slc35b2 | 14499 | -0.204 | -0.2135 | No |
| 60 | Atxn1 | 14855 | -0.222 | -0.2210 | Yes |
| 61 | Prdx5 | 14993 | -0.231 | -0.2170 | Yes |
| 62 | Bcl10 | 15014 | -0.232 | -0.2070 | Yes |
| 63 | Slc25a4 | 15032 | -0.233 | -0.1968 | Yes |
| 64 | Prdx1 | 15148 | -0.238 | -0.1913 | Yes |
| 65 | Pex13 | 15188 | -0.240 | -0.1819 | Yes |
| 66 | Slc25a17 | 15356 | -0.249 | -0.1785 | Yes |
| 67 | Cdk7 | 15408 | -0.252 | -0.1691 | Yes |
| 68 | Hsd17b11 | 15426 | -0.254 | -0.1579 | Yes |
| 69 | Fis1 | 15475 | -0.257 | -0.1481 | Yes |
| 70 | Pex2 | 15600 | -0.263 | -0.1419 | Yes |
| 71 | Hras | 16259 | -0.300 | -0.1611 | Yes |
| 72 | Sod1 | 16608 | -0.323 | -0.1634 | Yes |
| 73 | Sod2 | 16611 | -0.324 | -0.1481 | Yes |
| 74 | Mvp | 16923 | -0.341 | -0.1477 | Yes |
| 75 | Sult2b1 | 17208 | -0.362 | -0.1449 | Yes |
| 76 | Scp2 | 17345 | -0.374 | -0.1341 | Yes |
| 77 | Dlg4 | 17496 | -0.388 | -0.1233 | Yes |
| 78 | Hsd3b7 | 18072 | -0.437 | -0.1317 | Yes |
| 79 | Tspo | 18165 | -0.447 | -0.1152 | Yes |
| 80 | Acaa1a | 18247 | -0.455 | -0.0976 | Yes |
| 81 | Gstk1 | 18258 | -0.457 | -0.0764 | Yes |
| 82 | Cadm1 | 18356 | -0.470 | -0.0590 | Yes |
| 83 | Retsat | 18431 | -0.478 | -0.0400 | Yes |
| 84 | Abcb1a | 18549 | -0.494 | -0.0224 | Yes |
| 85 | Nudt19 | 18751 | -0.524 | -0.0077 | Yes |
| 86 | Pex11b | 19171 | -0.615 | 0.0002 | Yes |
| 87 | Eci2 | 19246 | -0.635 | 0.0267 | Yes |