Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group4.CMP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.22331403 |
| Normalized Enrichment Score (NES) | 0.8826489 |
| Nominal p-value | 0.754 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
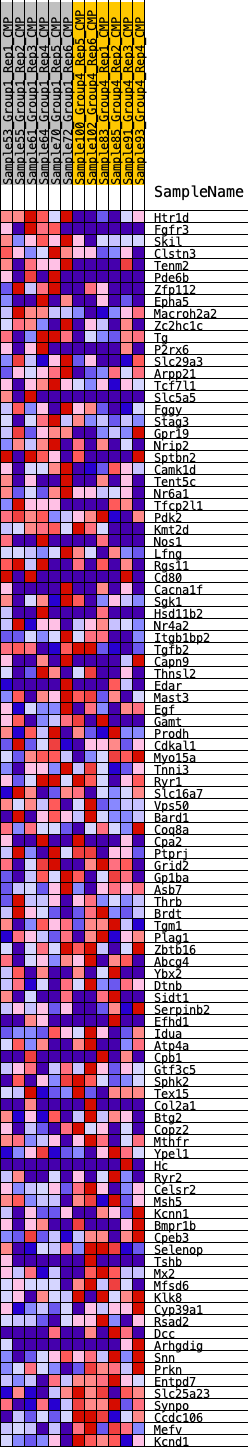
| 1 | Htr1d | 7 | 1.170 | 0.0449 | Yes |
| 2 | Fgfr3 | 127 | 0.772 | 0.0688 | Yes |
| 3 | Skil | 657 | 0.545 | 0.0630 | Yes |
| 4 | Clstn3 | 675 | 0.541 | 0.0830 | Yes |
| 5 | Tenm2 | 814 | 0.513 | 0.0959 | Yes |
| 6 | Pde6b | 920 | 0.497 | 0.1098 | Yes |
| 7 | Zfp112 | 1109 | 0.470 | 0.1184 | Yes |
| 8 | Epha5 | 1265 | 0.448 | 0.1279 | Yes |
| 9 | Macroh2a2 | 1273 | 0.448 | 0.1449 | Yes |
| 10 | Zc2hc1c | 1642 | 0.408 | 0.1420 | Yes |
| 11 | Tg | 1842 | 0.388 | 0.1468 | Yes |
| 12 | P2rx6 | 1972 | 0.376 | 0.1548 | Yes |
| 13 | Slc29a3 | 2330 | 0.349 | 0.1502 | Yes |
| 14 | Arpp21 | 2690 | 0.321 | 0.1444 | Yes |
| 15 | Tcf7l1 | 2864 | 0.310 | 0.1476 | Yes |
| 16 | Slc5a5 | 2886 | 0.308 | 0.1584 | Yes |
| 17 | Fggy | 3005 | 0.303 | 0.1642 | Yes |
| 18 | Stag3 | 3116 | 0.295 | 0.1700 | Yes |
| 19 | Gpr19 | 3252 | 0.287 | 0.1742 | Yes |
| 20 | Nrip2 | 3501 | 0.268 | 0.1720 | Yes |
| 21 | Sptbn2 | 3669 | 0.259 | 0.1735 | Yes |
| 22 | Camk1d | 3670 | 0.259 | 0.1835 | Yes |
| 23 | Tent5c | 3676 | 0.258 | 0.1933 | Yes |
| 24 | Nr6a1 | 3938 | 0.243 | 0.1894 | Yes |
| 25 | Tfcp2l1 | 4077 | 0.236 | 0.1915 | Yes |
| 26 | Pdk2 | 4129 | 0.233 | 0.1979 | Yes |
| 27 | Kmt2d | 4147 | 0.232 | 0.2061 | Yes |
| 28 | Nos1 | 4151 | 0.232 | 0.2149 | Yes |
| 29 | Lfng | 4162 | 0.231 | 0.2233 | Yes |
| 30 | Rgs11 | 4442 | 0.217 | 0.2175 | No |
| 31 | Cd80 | 5035 | 0.184 | 0.1946 | No |
| 32 | Cacna1f | 5438 | 0.168 | 0.1806 | No |
| 33 | Sgk1 | 5474 | 0.166 | 0.1853 | No |
| 34 | Hsd11b2 | 5905 | 0.145 | 0.1690 | No |
| 35 | Nr4a2 | 6757 | 0.113 | 0.1301 | No |
| 36 | Itgb1bp2 | 6902 | 0.107 | 0.1269 | No |
| 37 | Tgfb2 | 6905 | 0.106 | 0.1310 | No |
| 38 | Capn9 | 6934 | 0.105 | 0.1336 | No |
| 39 | Thnsl2 | 7256 | 0.091 | 0.1208 | No |
| 40 | Edar | 7395 | 0.085 | 0.1171 | No |
| 41 | Mast3 | 7552 | 0.079 | 0.1122 | No |
| 42 | Egf | 7570 | 0.078 | 0.1144 | No |
| 43 | Gamt | 7897 | 0.064 | 0.1003 | No |
| 44 | Prodh | 7945 | 0.062 | 0.1003 | No |
| 45 | Cdkal1 | 8271 | 0.049 | 0.0857 | No |
| 46 | Myo15a | 8331 | 0.047 | 0.0845 | No |
| 47 | Tnni3 | 8359 | 0.046 | 0.0849 | No |
| 48 | Ryr1 | 8370 | 0.045 | 0.0861 | No |
| 49 | Slc16a7 | 8585 | 0.036 | 0.0766 | No |
| 50 | Vps50 | 8646 | 0.034 | 0.0749 | No |
| 51 | Bard1 | 8720 | 0.031 | 0.0724 | No |
| 52 | Coq8a | 9318 | 0.008 | 0.0423 | No |
| 53 | Cpa2 | 9345 | 0.007 | 0.0413 | No |
| 54 | Ptprj | 9353 | 0.006 | 0.0412 | No |
| 55 | Grid2 | 9774 | -0.006 | 0.0200 | No |
| 56 | Gp1ba | 9824 | -0.008 | 0.0179 | No |
| 57 | Asb7 | 9845 | -0.009 | 0.0172 | No |
| 58 | Thrb | 9874 | -0.010 | 0.0162 | No |
| 59 | Brdt | 10054 | -0.016 | 0.0077 | No |
| 60 | Tgm1 | 10503 | -0.034 | -0.0138 | No |
| 61 | Plag1 | 11021 | -0.055 | -0.0379 | No |
| 62 | Zbtb16 | 11222 | -0.063 | -0.0457 | No |
| 63 | Abcg4 | 11761 | -0.086 | -0.0697 | No |
| 64 | Ybx2 | 11833 | -0.089 | -0.0699 | No |
| 65 | Dtnb | 11874 | -0.091 | -0.0684 | No |
| 66 | Sidt1 | 12076 | -0.100 | -0.0747 | No |
| 67 | Serpinb2 | 12216 | -0.106 | -0.0777 | No |
| 68 | Efhd1 | 12568 | -0.122 | -0.0908 | No |
| 69 | Idua | 12916 | -0.133 | -0.1033 | No |
| 70 | Atp4a | 12941 | -0.135 | -0.0993 | No |
| 71 | Cpb1 | 13192 | -0.146 | -0.1063 | No |
| 72 | Gtf3c5 | 13374 | -0.153 | -0.1096 | No |
| 73 | Sphk2 | 13564 | -0.162 | -0.1129 | No |
| 74 | Tex15 | 13737 | -0.169 | -0.1151 | No |
| 75 | Col2a1 | 13941 | -0.175 | -0.1187 | No |
| 76 | Btg2 | 14157 | -0.186 | -0.1224 | No |
| 77 | Copz2 | 14591 | -0.209 | -0.1364 | No |
| 78 | Mthfr | 14862 | -0.223 | -0.1415 | No |
| 79 | Ypel1 | 14982 | -0.230 | -0.1386 | No |
| 80 | Hc | 15238 | -0.242 | -0.1422 | No |
| 81 | Ryr2 | 15363 | -0.250 | -0.1388 | No |
| 82 | Celsr2 | 15418 | -0.253 | -0.1318 | No |
| 83 | Msh5 | 15703 | -0.268 | -0.1358 | No |
| 84 | Kcnn1 | 16215 | -0.297 | -0.1503 | No |
| 85 | Bmpr1b | 16234 | -0.298 | -0.1397 | No |
| 86 | Cpeb3 | 16337 | -0.306 | -0.1331 | No |
| 87 | Selenop | 16439 | -0.313 | -0.1261 | No |
| 88 | Tshb | 16703 | -0.329 | -0.1267 | No |
| 89 | Mx2 | 16802 | -0.333 | -0.1188 | No |
| 90 | Mfsd6 | 17273 | -0.368 | -0.1285 | No |
| 91 | Klk8 | 17340 | -0.373 | -0.1174 | No |
| 92 | Cyp39a1 | 17434 | -0.382 | -0.1073 | No |
| 93 | Rsad2 | 17533 | -0.389 | -0.0972 | No |
| 94 | Dcc | 17720 | -0.403 | -0.0911 | No |
| 95 | Arhgdig | 17837 | -0.412 | -0.0810 | No |
| 96 | Snn | 17989 | -0.428 | -0.0721 | No |
| 97 | Prkn | 18126 | -0.443 | -0.0619 | No |
| 98 | Entpd7 | 18480 | -0.485 | -0.0611 | No |
| 99 | Slc25a23 | 18822 | -0.538 | -0.0576 | No |
| 100 | Synpo | 19128 | -0.604 | -0.0497 | No |
| 101 | Ccdc106 | 19309 | -0.653 | -0.0336 | No |
| 102 | Mefv | 19496 | -0.725 | -0.0150 | No |
| 103 | Kcnd1 | 19535 | -0.746 | 0.0120 | No |