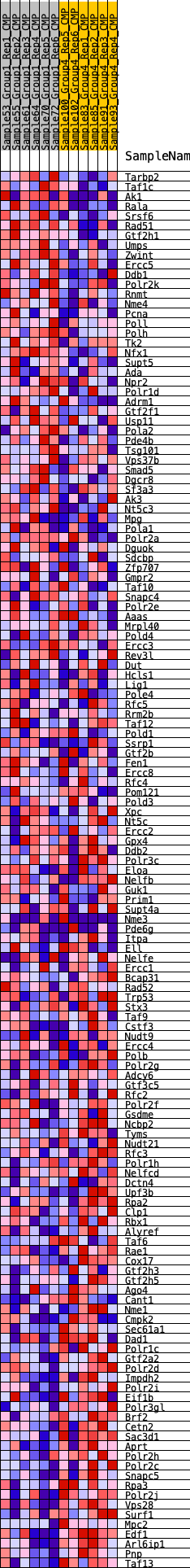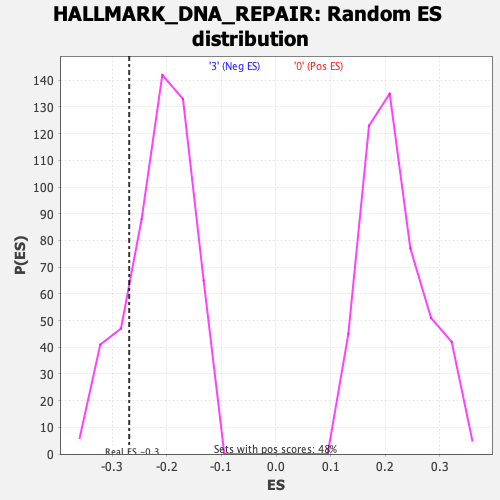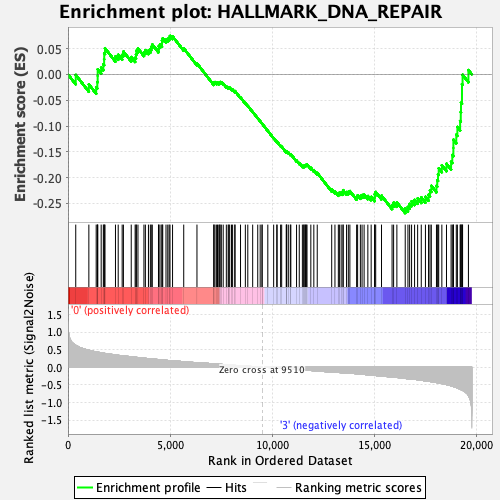
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group4.CMP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_DNA_REPAIR |
| Enrichment Score (ES) | -0.26881558 |
| Normalized Enrichment Score (NES) | -1.2644188 |
| Nominal p-value | 0.1724138 |
| FDR q-value | 0.3412618 |
| FWER p-Value | 0.81 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tarbp2 | 379 | 0.621 | -0.0009 | No |
| 2 | Taf1c | 1021 | 0.484 | -0.0193 | No |
| 3 | Ak1 | 1380 | 0.435 | -0.0246 | No |
| 4 | Rala | 1435 | 0.430 | -0.0147 | No |
| 5 | Srsf6 | 1452 | 0.429 | -0.0028 | No |
| 6 | Rad51 | 1454 | 0.429 | 0.0098 | No |
| 7 | Gtf2h1 | 1624 | 0.409 | 0.0133 | No |
| 8 | Umps | 1730 | 0.399 | 0.0198 | No |
| 9 | Zwint | 1769 | 0.395 | 0.0295 | No |
| 10 | Ercc5 | 1774 | 0.395 | 0.0410 | No |
| 11 | Ddb1 | 1811 | 0.391 | 0.0508 | No |
| 12 | Polr2k | 2324 | 0.350 | 0.0350 | No |
| 13 | Rnmt | 2458 | 0.338 | 0.0383 | No |
| 14 | Nme4 | 2652 | 0.325 | 0.0380 | No |
| 15 | Pcna | 2713 | 0.319 | 0.0444 | No |
| 16 | Poll | 3096 | 0.296 | 0.0337 | No |
| 17 | Polh | 3287 | 0.285 | 0.0324 | No |
| 18 | Tk2 | 3330 | 0.282 | 0.0386 | No |
| 19 | Nfx1 | 3347 | 0.280 | 0.0461 | No |
| 20 | Supt5 | 3425 | 0.274 | 0.0503 | No |
| 21 | Ada | 3718 | 0.256 | 0.0430 | No |
| 22 | Npr2 | 3788 | 0.254 | 0.0470 | No |
| 23 | Polr1d | 3939 | 0.243 | 0.0465 | No |
| 24 | Adrm1 | 4031 | 0.239 | 0.0490 | No |
| 25 | Gtf2f1 | 4076 | 0.236 | 0.0537 | No |
| 26 | Usp11 | 4127 | 0.233 | 0.0580 | No |
| 27 | Pola2 | 4430 | 0.218 | 0.0491 | No |
| 28 | Pde4b | 4445 | 0.217 | 0.0548 | No |
| 29 | Tsg101 | 4504 | 0.214 | 0.0582 | No |
| 30 | Vps37b | 4601 | 0.208 | 0.0595 | No |
| 31 | Smad5 | 4602 | 0.208 | 0.0656 | No |
| 32 | Dgcr8 | 4638 | 0.207 | 0.0700 | No |
| 33 | Sf3a3 | 4798 | 0.198 | 0.0677 | No |
| 34 | Ak3 | 4885 | 0.193 | 0.0690 | No |
| 35 | Nt5c3 | 4951 | 0.189 | 0.0713 | No |
| 36 | Mpg | 4994 | 0.187 | 0.0747 | No |
| 37 | Pola1 | 5119 | 0.180 | 0.0737 | No |
| 38 | Polr2a | 5666 | 0.158 | 0.0506 | No |
| 39 | Dguok | 6314 | 0.128 | 0.0214 | No |
| 40 | Sdcbp | 7138 | 0.097 | -0.0177 | No |
| 41 | Zfp707 | 7143 | 0.097 | -0.0150 | No |
| 42 | Gmpr2 | 7196 | 0.094 | -0.0149 | No |
| 43 | Taf10 | 7277 | 0.090 | -0.0163 | No |
| 44 | Snapc4 | 7320 | 0.089 | -0.0158 | No |
| 45 | Polr2e | 7385 | 0.086 | -0.0165 | No |
| 46 | Aaas | 7428 | 0.084 | -0.0162 | No |
| 47 | Mrpl40 | 7447 | 0.083 | -0.0147 | No |
| 48 | Pold4 | 7507 | 0.081 | -0.0153 | No |
| 49 | Ercc3 | 7599 | 0.077 | -0.0176 | No |
| 50 | Rev3l | 7752 | 0.071 | -0.0233 | No |
| 51 | Dut | 7841 | 0.066 | -0.0258 | No |
| 52 | Hcls1 | 7872 | 0.065 | -0.0254 | No |
| 53 | Lig1 | 7916 | 0.063 | -0.0257 | No |
| 54 | Pole4 | 8004 | 0.059 | -0.0284 | No |
| 55 | Rfc5 | 8059 | 0.057 | -0.0295 | No |
| 56 | Rrm2b | 8166 | 0.052 | -0.0333 | No |
| 57 | Taf12 | 8178 | 0.052 | -0.0323 | No |
| 58 | Pold1 | 8447 | 0.042 | -0.0447 | No |
| 59 | Ssrp1 | 8683 | 0.033 | -0.0557 | No |
| 60 | Gtf2b | 8803 | 0.028 | -0.0610 | No |
| 61 | Fen1 | 9042 | 0.019 | -0.0725 | No |
| 62 | Ercc8 | 9290 | 0.009 | -0.0849 | No |
| 63 | Rfc4 | 9409 | 0.004 | -0.0908 | No |
| 64 | Pom121 | 9501 | 0.000 | -0.0954 | No |
| 65 | Pold3 | 9503 | 0.000 | -0.0954 | No |
| 66 | Xpc | 9783 | -0.006 | -0.1095 | No |
| 67 | Nt5c | 10069 | -0.017 | -0.1235 | No |
| 68 | Ercc2 | 10222 | -0.023 | -0.1305 | No |
| 69 | Gpx4 | 10238 | -0.024 | -0.1306 | No |
| 70 | Ddb2 | 10409 | -0.030 | -0.1384 | No |
| 71 | Polr3c | 10462 | -0.032 | -0.1401 | No |
| 72 | Eloa | 10685 | -0.041 | -0.1502 | No |
| 73 | Nelfb | 10700 | -0.042 | -0.1497 | No |
| 74 | Guk1 | 10708 | -0.042 | -0.1488 | No |
| 75 | Prim1 | 10787 | -0.045 | -0.1514 | No |
| 76 | Supt4a | 10884 | -0.050 | -0.1548 | No |
| 77 | Nme3 | 10910 | -0.050 | -0.1546 | No |
| 78 | Pde6g | 11190 | -0.062 | -0.1670 | No |
| 79 | Itpa | 11323 | -0.068 | -0.1717 | No |
| 80 | Ell | 11475 | -0.074 | -0.1772 | No |
| 81 | Nelfe | 11541 | -0.077 | -0.1782 | No |
| 82 | Ercc1 | 11556 | -0.078 | -0.1766 | No |
| 83 | Bcap31 | 11603 | -0.080 | -0.1766 | No |
| 84 | Rad52 | 11631 | -0.081 | -0.1756 | No |
| 85 | Trp53 | 11669 | -0.082 | -0.1750 | No |
| 86 | Stx3 | 11702 | -0.084 | -0.1742 | No |
| 87 | Taf9 | 11889 | -0.092 | -0.1810 | No |
| 88 | Cstf3 | 12036 | -0.098 | -0.1855 | No |
| 89 | Nudt9 | 12203 | -0.105 | -0.1909 | No |
| 90 | Ercc4 | 12911 | -0.133 | -0.2229 | No |
| 91 | Polb | 13070 | -0.141 | -0.2268 | No |
| 92 | Polr2g | 13236 | -0.148 | -0.2309 | No |
| 93 | Adcy6 | 13287 | -0.150 | -0.2290 | No |
| 94 | Gtf3c5 | 13374 | -0.153 | -0.2288 | No |
| 95 | Rfc2 | 13464 | -0.158 | -0.2287 | No |
| 96 | Polr2f | 13471 | -0.158 | -0.2243 | No |
| 97 | Gsdme | 13638 | -0.165 | -0.2279 | No |
| 98 | Ncbp2 | 13725 | -0.169 | -0.2273 | No |
| 99 | Tyms | 13801 | -0.172 | -0.2260 | No |
| 100 | Nudt21 | 14131 | -0.184 | -0.2373 | No |
| 101 | Rfc3 | 14180 | -0.187 | -0.2343 | No |
| 102 | Polr1h | 14315 | -0.194 | -0.2354 | No |
| 103 | Nelfcd | 14397 | -0.198 | -0.2336 | No |
| 104 | Dctn4 | 14498 | -0.204 | -0.2327 | No |
| 105 | Upf3b | 14681 | -0.213 | -0.2356 | No |
| 106 | Rpa2 | 14843 | -0.222 | -0.2373 | No |
| 107 | Clp1 | 15013 | -0.232 | -0.2390 | No |
| 108 | Rbx1 | 15016 | -0.232 | -0.2323 | No |
| 109 | Alyref | 15065 | -0.234 | -0.2278 | No |
| 110 | Taf6 | 15350 | -0.249 | -0.2349 | No |
| 111 | Rae1 | 15867 | -0.275 | -0.2530 | No |
| 112 | Cox17 | 15942 | -0.280 | -0.2485 | No |
| 113 | Gtf2h3 | 16102 | -0.289 | -0.2480 | No |
| 114 | Gtf2h5 | 16511 | -0.319 | -0.2594 | Yes |
| 115 | Ago4 | 16644 | -0.325 | -0.2565 | Yes |
| 116 | Cant1 | 16727 | -0.330 | -0.2509 | Yes |
| 117 | Nme1 | 16822 | -0.334 | -0.2458 | Yes |
| 118 | Cmpk2 | 16969 | -0.342 | -0.2432 | Yes |
| 119 | Sec61a1 | 17119 | -0.355 | -0.2402 | Yes |
| 120 | Dad1 | 17296 | -0.370 | -0.2383 | Yes |
| 121 | Polr1c | 17500 | -0.388 | -0.2371 | Yes |
| 122 | Gtf2a2 | 17653 | -0.399 | -0.2330 | Yes |
| 123 | Polr2d | 17723 | -0.404 | -0.2246 | Yes |
| 124 | Impdh2 | 17788 | -0.410 | -0.2157 | Yes |
| 125 | Polr2i | 18040 | -0.434 | -0.2157 | Yes |
| 126 | Eif1b | 18079 | -0.437 | -0.2047 | Yes |
| 127 | Polr3gl | 18117 | -0.442 | -0.1935 | Yes |
| 128 | Brf2 | 18151 | -0.445 | -0.1820 | Yes |
| 129 | Cetn2 | 18304 | -0.463 | -0.1760 | Yes |
| 130 | Sac3d1 | 18531 | -0.491 | -0.1730 | Yes |
| 131 | Aprt | 18756 | -0.525 | -0.1689 | Yes |
| 132 | Polr2h | 18815 | -0.536 | -0.1560 | Yes |
| 133 | Polr2c | 18868 | -0.546 | -0.1425 | Yes |
| 134 | Snapc5 | 18870 | -0.546 | -0.1264 | Yes |
| 135 | Rpa3 | 19011 | -0.575 | -0.1165 | Yes |
| 136 | Polr2j | 19065 | -0.590 | -0.1018 | Yes |
| 137 | Vps28 | 19199 | -0.623 | -0.0901 | Yes |
| 138 | Surf1 | 19231 | -0.632 | -0.0730 | Yes |
| 139 | Mpc2 | 19242 | -0.635 | -0.0547 | Yes |
| 140 | Edf1 | 19290 | -0.648 | -0.0379 | Yes |
| 141 | Arl6ip1 | 19291 | -0.649 | -0.0188 | Yes |
| 142 | Pnp | 19319 | -0.657 | -0.0007 | Yes |
| 143 | Taf13 | 19605 | -0.800 | 0.0085 | Yes |