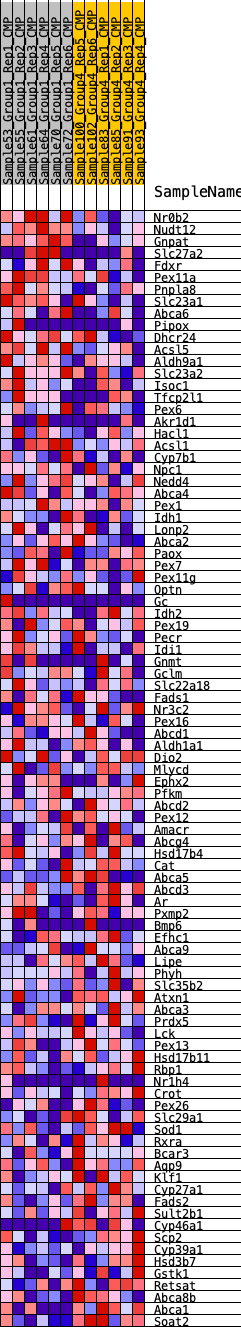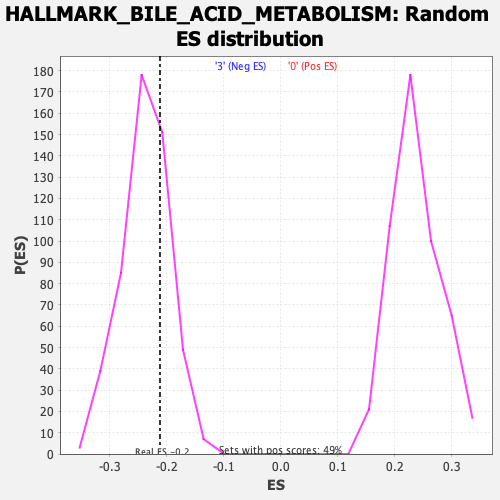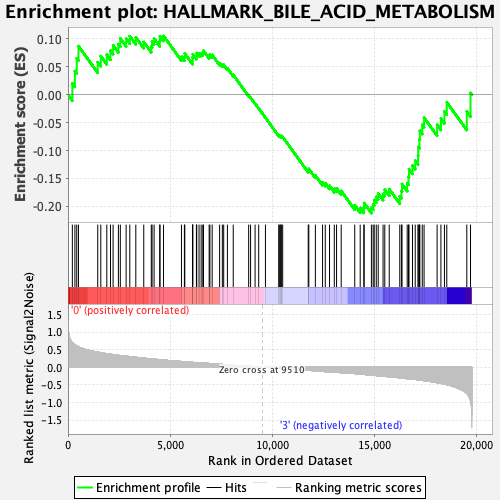
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group4.CMP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.21221209 |
| Normalized Enrichment Score (NES) | -0.894318 |
| Nominal p-value | 0.7265625 |
| FDR q-value | 0.7374457 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nr0b2 | 213 | 0.693 | 0.0203 | No |
| 2 | Nudt12 | 339 | 0.636 | 0.0425 | No |
| 3 | Gnpat | 428 | 0.607 | 0.0652 | No |
| 4 | Slc27a2 | 512 | 0.581 | 0.0871 | No |
| 5 | Fdxr | 1456 | 0.428 | 0.0584 | No |
| 6 | Pex11a | 1606 | 0.411 | 0.0693 | No |
| 7 | Pnpla8 | 1900 | 0.383 | 0.0716 | No |
| 8 | Slc23a1 | 2080 | 0.368 | 0.0791 | No |
| 9 | Abca6 | 2208 | 0.359 | 0.0887 | No |
| 10 | Pipox | 2467 | 0.338 | 0.0908 | No |
| 11 | Dhcr24 | 2562 | 0.331 | 0.1009 | No |
| 12 | Acsl5 | 2846 | 0.312 | 0.1005 | No |
| 13 | Aldh9a1 | 3025 | 0.302 | 0.1050 | No |
| 14 | Slc23a2 | 3320 | 0.283 | 0.1028 | No |
| 15 | Isoc1 | 3709 | 0.257 | 0.0946 | No |
| 16 | Tfcp2l1 | 4077 | 0.236 | 0.0865 | No |
| 17 | Pex6 | 4116 | 0.234 | 0.0951 | No |
| 18 | Akr1d1 | 4215 | 0.228 | 0.1004 | No |
| 19 | Hacl1 | 4493 | 0.214 | 0.0959 | No |
| 20 | Acsl1 | 4503 | 0.214 | 0.1051 | No |
| 21 | Cyp7b1 | 4677 | 0.204 | 0.1054 | No |
| 22 | Npc1 | 5555 | 0.163 | 0.0682 | No |
| 23 | Nedd4 | 5699 | 0.155 | 0.0679 | No |
| 24 | Abca4 | 5718 | 0.154 | 0.0739 | No |
| 25 | Pex1 | 6097 | 0.137 | 0.0608 | No |
| 26 | Idh1 | 6098 | 0.137 | 0.0670 | No |
| 27 | Lonp2 | 6112 | 0.136 | 0.0725 | No |
| 28 | Abca2 | 6287 | 0.129 | 0.0694 | No |
| 29 | Paox | 6303 | 0.128 | 0.0744 | No |
| 30 | Pex7 | 6416 | 0.123 | 0.0742 | No |
| 31 | Pex11g | 6519 | 0.118 | 0.0743 | No |
| 32 | Optn | 6600 | 0.114 | 0.0754 | No |
| 33 | Gc | 6634 | 0.114 | 0.0788 | No |
| 34 | Idh2 | 6911 | 0.106 | 0.0696 | No |
| 35 | Pex19 | 6948 | 0.104 | 0.0724 | No |
| 36 | Pecr | 7059 | 0.099 | 0.0713 | No |
| 37 | Idi1 | 7423 | 0.084 | 0.0566 | No |
| 38 | Gnmt | 7557 | 0.079 | 0.0534 | No |
| 39 | Gclm | 7623 | 0.076 | 0.0535 | No |
| 40 | Slc22a18 | 7805 | 0.068 | 0.0474 | No |
| 41 | Fads1 | 8090 | 0.056 | 0.0355 | No |
| 42 | Nr3c2 | 8845 | 0.026 | -0.0016 | No |
| 43 | Pex16 | 8939 | 0.023 | -0.0053 | No |
| 44 | Abcd1 | 9160 | 0.013 | -0.0159 | No |
| 45 | Aldh1a1 | 9337 | 0.007 | -0.0245 | No |
| 46 | Dio2 | 9667 | -0.002 | -0.0412 | No |
| 47 | Mlycd | 10318 | -0.026 | -0.0730 | No |
| 48 | Ephx2 | 10348 | -0.027 | -0.0733 | No |
| 49 | Pfkm | 10416 | -0.030 | -0.0753 | No |
| 50 | Abcd2 | 10429 | -0.031 | -0.0745 | No |
| 51 | Pex12 | 10440 | -0.031 | -0.0736 | No |
| 52 | Amacr | 10511 | -0.034 | -0.0757 | No |
| 53 | Abcg4 | 11761 | -0.086 | -0.1353 | No |
| 54 | Hsd17b4 | 11791 | -0.087 | -0.1329 | No |
| 55 | Cat | 12111 | -0.101 | -0.1446 | No |
| 56 | Abca5 | 12463 | -0.117 | -0.1571 | No |
| 57 | Abcd3 | 12596 | -0.123 | -0.1583 | No |
| 58 | Ar | 12805 | -0.128 | -0.1631 | No |
| 59 | Pxmp2 | 13038 | -0.139 | -0.1687 | No |
| 60 | Bmp6 | 13149 | -0.144 | -0.1678 | No |
| 61 | Efhc1 | 13376 | -0.153 | -0.1724 | No |
| 62 | Abca9 | 14039 | -0.179 | -0.1980 | No |
| 63 | Lipe | 14306 | -0.193 | -0.2028 | Yes |
| 64 | Phyh | 14486 | -0.204 | -0.2027 | Yes |
| 65 | Slc35b2 | 14499 | -0.204 | -0.1942 | Yes |
| 66 | Atxn1 | 14855 | -0.222 | -0.2022 | Yes |
| 67 | Abca3 | 14940 | -0.227 | -0.1963 | Yes |
| 68 | Prdx5 | 14993 | -0.231 | -0.1886 | Yes |
| 69 | Lck | 15093 | -0.236 | -0.1830 | Yes |
| 70 | Pex13 | 15188 | -0.240 | -0.1770 | Yes |
| 71 | Hsd17b11 | 15426 | -0.254 | -0.1777 | Yes |
| 72 | Rbp1 | 15510 | -0.258 | -0.1703 | Yes |
| 73 | Nr1h4 | 15728 | -0.270 | -0.1692 | Yes |
| 74 | Crot | 16244 | -0.299 | -0.1820 | Yes |
| 75 | Pex26 | 16324 | -0.305 | -0.1723 | Yes |
| 76 | Slc29a1 | 16354 | -0.307 | -0.1600 | Yes |
| 77 | Sod1 | 16608 | -0.323 | -0.1583 | Yes |
| 78 | Rxra | 16671 | -0.326 | -0.1468 | Yes |
| 79 | Bcar3 | 16696 | -0.328 | -0.1333 | Yes |
| 80 | Aqp9 | 16867 | -0.336 | -0.1268 | Yes |
| 81 | Klf1 | 17000 | -0.345 | -0.1180 | Yes |
| 82 | Cyp27a1 | 17140 | -0.357 | -0.1091 | Yes |
| 83 | Fads2 | 17148 | -0.357 | -0.0934 | Yes |
| 84 | Sult2b1 | 17208 | -0.362 | -0.0801 | Yes |
| 85 | Cyp46a1 | 17227 | -0.364 | -0.0647 | Yes |
| 86 | Scp2 | 17345 | -0.374 | -0.0538 | Yes |
| 87 | Cyp39a1 | 17434 | -0.382 | -0.0411 | Yes |
| 88 | Hsd3b7 | 18072 | -0.437 | -0.0539 | Yes |
| 89 | Gstk1 | 18258 | -0.457 | -0.0428 | Yes |
| 90 | Retsat | 18431 | -0.478 | -0.0300 | Yes |
| 91 | Abca8b | 18546 | -0.494 | -0.0136 | Yes |
| 92 | Abca1 | 19526 | -0.742 | -0.0301 | Yes |
| 93 | Soat2 | 19708 | -0.945 | 0.0032 | Yes |