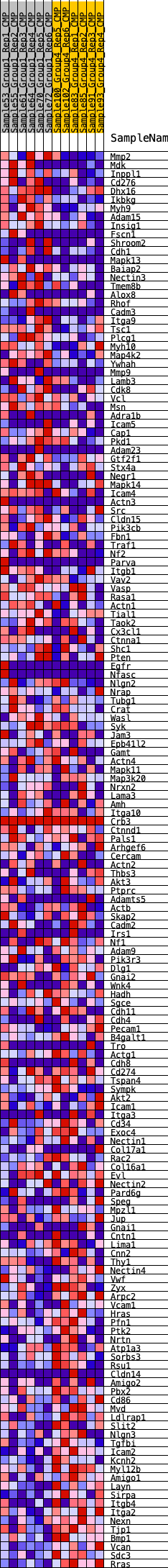
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group4.CMP_Pheno.cls#Group1_versus_Group4_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
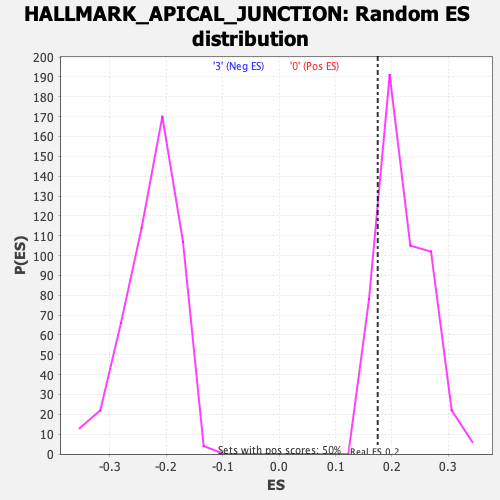
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | 0.17477672 |
| Normalized Enrichment Score (NES) | 0.7914844 |
| Nominal p-value | 0.87103176 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mmp2 | 135 | 0.759 | 0.0110 | Yes |
| 2 | Mdk | 144 | 0.752 | 0.0283 | Yes |
| 3 | Inppl1 | 168 | 0.722 | 0.0442 | Yes |
| 4 | Cd276 | 261 | 0.669 | 0.0552 | Yes |
| 5 | Dhx16 | 469 | 0.594 | 0.0587 | Yes |
| 6 | Ikbkg | 542 | 0.572 | 0.0685 | Yes |
| 7 | Myh9 | 568 | 0.562 | 0.0805 | Yes |
| 8 | Adam15 | 819 | 0.512 | 0.0798 | Yes |
| 9 | Insig1 | 906 | 0.499 | 0.0872 | Yes |
| 10 | Fscn1 | 1143 | 0.465 | 0.0861 | Yes |
| 11 | Shroom2 | 1148 | 0.464 | 0.0968 | Yes |
| 12 | Cdh1 | 1242 | 0.452 | 0.1027 | Yes |
| 13 | Mapk13 | 1335 | 0.441 | 0.1084 | Yes |
| 14 | Baiap2 | 1384 | 0.435 | 0.1162 | Yes |
| 15 | Nectin3 | 1506 | 0.423 | 0.1200 | Yes |
| 16 | Tmem8b | 1545 | 0.418 | 0.1279 | Yes |
| 17 | Alox8 | 1591 | 0.412 | 0.1354 | Yes |
| 18 | Rhof | 1622 | 0.409 | 0.1435 | Yes |
| 19 | Cadm3 | 1883 | 0.384 | 0.1393 | Yes |
| 20 | Itga9 | 1947 | 0.378 | 0.1450 | Yes |
| 21 | Tsc1 | 2133 | 0.366 | 0.1442 | Yes |
| 22 | Plcg1 | 2289 | 0.352 | 0.1446 | Yes |
| 23 | Myh10 | 2491 | 0.336 | 0.1422 | Yes |
| 24 | Map4k2 | 2527 | 0.333 | 0.1483 | Yes |
| 25 | Ywhah | 2586 | 0.328 | 0.1531 | Yes |
| 26 | Mmp9 | 2618 | 0.325 | 0.1592 | Yes |
| 27 | Lamb3 | 2764 | 0.318 | 0.1593 | Yes |
| 28 | Cdk8 | 2771 | 0.318 | 0.1664 | Yes |
| 29 | Vcl | 2938 | 0.307 | 0.1652 | Yes |
| 30 | Msn | 3131 | 0.293 | 0.1623 | Yes |
| 31 | Adra1b | 3228 | 0.288 | 0.1642 | Yes |
| 32 | Icam5 | 3286 | 0.285 | 0.1680 | Yes |
| 33 | Cap1 | 3300 | 0.284 | 0.1741 | Yes |
| 34 | Pkd1 | 3465 | 0.271 | 0.1721 | Yes |
| 35 | Adam23 | 3536 | 0.266 | 0.1748 | Yes |
| 36 | Gtf2f1 | 4076 | 0.236 | 0.1529 | No |
| 37 | Stx4a | 4191 | 0.229 | 0.1524 | No |
| 38 | Negr1 | 4271 | 0.226 | 0.1537 | No |
| 39 | Mapk14 | 4339 | 0.222 | 0.1556 | No |
| 40 | Icam4 | 4355 | 0.221 | 0.1600 | No |
| 41 | Actn3 | 4398 | 0.219 | 0.1630 | No |
| 42 | Src | 4611 | 0.208 | 0.1571 | No |
| 43 | Cldn15 | 4669 | 0.205 | 0.1590 | No |
| 44 | Pik3cb | 4744 | 0.201 | 0.1600 | No |
| 45 | Fbn1 | 4819 | 0.196 | 0.1608 | No |
| 46 | Traf1 | 4943 | 0.190 | 0.1590 | No |
| 47 | Nf2 | 5151 | 0.179 | 0.1527 | No |
| 48 | Parva | 5186 | 0.178 | 0.1551 | No |
| 49 | Itgb1 | 5328 | 0.172 | 0.1520 | No |
| 50 | Vav2 | 5332 | 0.172 | 0.1559 | No |
| 51 | Vasp | 5385 | 0.170 | 0.1573 | No |
| 52 | Rasa1 | 5391 | 0.169 | 0.1610 | No |
| 53 | Actn1 | 5544 | 0.163 | 0.1571 | No |
| 54 | Tial1 | 5945 | 0.144 | 0.1401 | No |
| 55 | Taok2 | 6056 | 0.139 | 0.1377 | No |
| 56 | Cx3cl1 | 6116 | 0.136 | 0.1380 | No |
| 57 | Ctnna1 | 6467 | 0.120 | 0.1229 | No |
| 58 | Shc1 | 6502 | 0.119 | 0.1240 | No |
| 59 | Pten | 6552 | 0.117 | 0.1243 | No |
| 60 | Egfr | 6620 | 0.114 | 0.1235 | No |
| 61 | Nfasc | 6625 | 0.114 | 0.1260 | No |
| 62 | Nlgn2 | 6984 | 0.102 | 0.1101 | No |
| 63 | Nrap | 7250 | 0.092 | 0.0988 | No |
| 64 | Tubg1 | 7275 | 0.090 | 0.0997 | No |
| 65 | Crat | 7440 | 0.083 | 0.0933 | No |
| 66 | Wasl | 7469 | 0.082 | 0.0938 | No |
| 67 | Syk | 7548 | 0.079 | 0.0917 | No |
| 68 | Jam3 | 7602 | 0.077 | 0.0908 | No |
| 69 | Epb41l2 | 7644 | 0.076 | 0.0905 | No |
| 70 | Gamt | 7897 | 0.064 | 0.0792 | No |
| 71 | Actn4 | 8301 | 0.048 | 0.0597 | No |
| 72 | Mapk11 | 8537 | 0.039 | 0.0486 | No |
| 73 | Map3k20 | 8635 | 0.035 | 0.0445 | No |
| 74 | Nrxn2 | 8680 | 0.033 | 0.0430 | No |
| 75 | Lama3 | 9013 | 0.021 | 0.0266 | No |
| 76 | Amh | 9277 | 0.009 | 0.0134 | No |
| 77 | Itga10 | 9348 | 0.007 | 0.0100 | No |
| 78 | Crb3 | 9547 | 0.000 | -0.0001 | No |
| 79 | Ctnnd1 | 9811 | -0.008 | -0.0133 | No |
| 80 | Pals1 | 9860 | -0.009 | -0.0156 | No |
| 81 | Arhgef6 | 9917 | -0.011 | -0.0181 | No |
| 82 | Cercam | 10057 | -0.016 | -0.0249 | No |
| 83 | Actn2 | 10460 | -0.032 | -0.0446 | No |
| 84 | Thbs3 | 10488 | -0.033 | -0.0452 | No |
| 85 | Akt3 | 10509 | -0.034 | -0.0454 | No |
| 86 | Ptprc | 10539 | -0.035 | -0.0461 | No |
| 87 | Adamts5 | 10728 | -0.043 | -0.0546 | No |
| 88 | Actb | 10748 | -0.044 | -0.0546 | No |
| 89 | Skap2 | 10768 | -0.045 | -0.0545 | No |
| 90 | Cadm2 | 11024 | -0.055 | -0.0662 | No |
| 91 | Irs1 | 11054 | -0.056 | -0.0664 | No |
| 92 | Nf1 | 11252 | -0.065 | -0.0749 | No |
| 93 | Adam9 | 11257 | -0.065 | -0.0736 | No |
| 94 | Pik3r3 | 11346 | -0.068 | -0.0764 | No |
| 95 | Dlg1 | 11412 | -0.071 | -0.0781 | No |
| 96 | Gnai2 | 11481 | -0.074 | -0.0798 | No |
| 97 | Wnk4 | 11544 | -0.078 | -0.0811 | No |
| 98 | Hadh | 11709 | -0.084 | -0.0875 | No |
| 99 | Sgce | 12109 | -0.100 | -0.1055 | No |
| 100 | Cdh11 | 12166 | -0.103 | -0.1059 | No |
| 101 | Cdh4 | 12487 | -0.118 | -0.1194 | No |
| 102 | Pecam1 | 12547 | -0.121 | -0.1196 | No |
| 103 | B4galt1 | 12598 | -0.123 | -0.1192 | No |
| 104 | Tro | 12685 | -0.127 | -0.1206 | No |
| 105 | Actg1 | 12783 | -0.127 | -0.1226 | No |
| 106 | Cdh8 | 12807 | -0.128 | -0.1207 | No |
| 107 | Cd274 | 12814 | -0.128 | -0.1180 | No |
| 108 | Tspan4 | 13006 | -0.138 | -0.1245 | No |
| 109 | Sympk | 13030 | -0.139 | -0.1224 | No |
| 110 | Akt2 | 13141 | -0.144 | -0.1246 | No |
| 111 | Icam1 | 13290 | -0.150 | -0.1286 | No |
| 112 | Itga3 | 13315 | -0.151 | -0.1263 | No |
| 113 | Cd34 | 13354 | -0.152 | -0.1247 | No |
| 114 | Exoc4 | 13360 | -0.153 | -0.1213 | No |
| 115 | Nectin1 | 13472 | -0.158 | -0.1232 | No |
| 116 | Col17a1 | 13606 | -0.164 | -0.1262 | No |
| 117 | Rac2 | 13747 | -0.170 | -0.1293 | No |
| 118 | Col16a1 | 13953 | -0.175 | -0.1356 | No |
| 119 | Evl | 14223 | -0.189 | -0.1449 | No |
| 120 | Nectin2 | 14299 | -0.193 | -0.1442 | No |
| 121 | Pard6g | 14342 | -0.195 | -0.1417 | No |
| 122 | Speg | 14699 | -0.215 | -0.1548 | No |
| 123 | Mpzl1 | 14719 | -0.215 | -0.1507 | No |
| 124 | Jup | 14794 | -0.220 | -0.1493 | No |
| 125 | Gnai1 | 14841 | -0.222 | -0.1464 | No |
| 126 | Cntn1 | 14934 | -0.227 | -0.1458 | No |
| 127 | Lima1 | 15169 | -0.239 | -0.1521 | No |
| 128 | Cnn2 | 15362 | -0.250 | -0.1560 | No |
| 129 | Thy1 | 15433 | -0.254 | -0.1535 | No |
| 130 | Nectin4 | 15598 | -0.263 | -0.1557 | No |
| 131 | Vwf | 15776 | -0.271 | -0.1584 | No |
| 132 | Zyx | 15893 | -0.277 | -0.1577 | No |
| 133 | Arpc2 | 16095 | -0.289 | -0.1612 | No |
| 134 | Vcam1 | 16258 | -0.300 | -0.1624 | No |
| 135 | Hras | 16259 | -0.300 | -0.1553 | No |
| 136 | Pfn1 | 16266 | -0.300 | -0.1486 | No |
| 137 | Ptk2 | 16325 | -0.305 | -0.1443 | No |
| 138 | Nrtn | 16400 | -0.311 | -0.1408 | No |
| 139 | Atp1a3 | 16482 | -0.317 | -0.1374 | No |
| 140 | Sorbs3 | 16503 | -0.318 | -0.1310 | No |
| 141 | Rsu1 | 16550 | -0.320 | -0.1258 | No |
| 142 | Cldn14 | 16597 | -0.323 | -0.1205 | No |
| 143 | Amigo2 | 16661 | -0.325 | -0.1160 | No |
| 144 | Pbx2 | 16813 | -0.334 | -0.1159 | No |
| 145 | Cd86 | 17085 | -0.352 | -0.1214 | No |
| 146 | Mvd | 17161 | -0.359 | -0.1168 | No |
| 147 | Ldlrap1 | 17191 | -0.361 | -0.1097 | No |
| 148 | Slit2 | 17356 | -0.374 | -0.1093 | No |
| 149 | Nlgn3 | 17476 | -0.386 | -0.1062 | No |
| 150 | Tgfbi | 17888 | -0.418 | -0.1174 | No |
| 151 | Icam2 | 18421 | -0.477 | -0.1332 | No |
| 152 | Kcnh2 | 18472 | -0.483 | -0.1244 | No |
| 153 | Myl12b | 18805 | -0.534 | -0.1288 | No |
| 154 | Amigo1 | 18840 | -0.540 | -0.1178 | No |
| 155 | Layn | 18985 | -0.570 | -0.1117 | No |
| 156 | Sirpa | 19113 | -0.601 | -0.1040 | No |
| 157 | Itgb4 | 19146 | -0.609 | -0.0913 | No |
| 158 | Itga2 | 19222 | -0.629 | -0.0803 | No |
| 159 | Nexn | 19230 | -0.631 | -0.0657 | No |
| 160 | Tjp1 | 19263 | -0.640 | -0.0523 | No |
| 161 | Bmp1 | 19312 | -0.655 | -0.0393 | No |
| 162 | Vcan | 19495 | -0.724 | -0.0315 | No |
| 163 | Sdc3 | 19498 | -0.727 | -0.0145 | No |
| 164 | Rras | 19757 | -1.203 | 0.0007 | No |