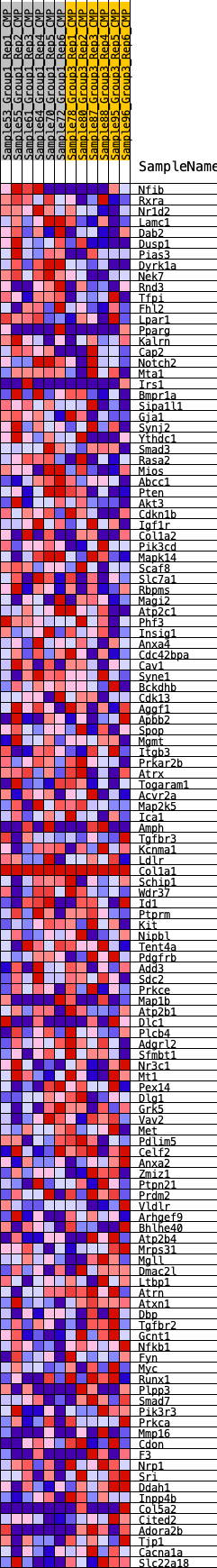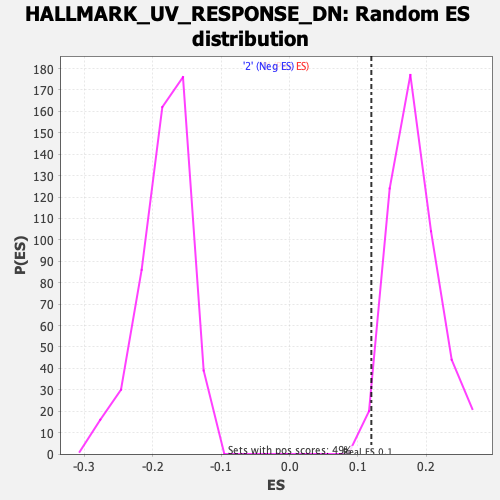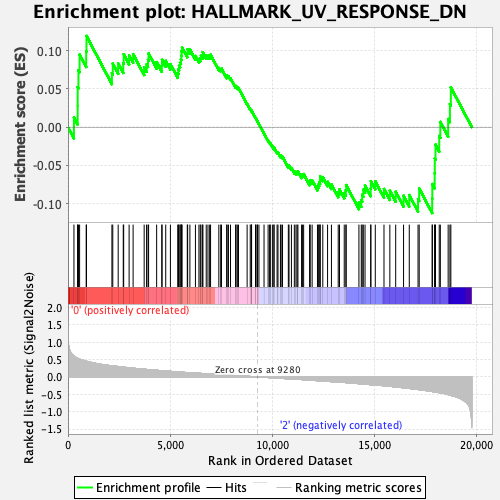
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group3.CMP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.119518094 |
| Normalized Enrichment Score (NES) | 0.656839 |
| Nominal p-value | 0.9959184 |
| FDR q-value | 0.9973094 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nfib | 291 | 0.601 | 0.0127 | Yes |
| 2 | Rxra | 473 | 0.535 | 0.0280 | Yes |
| 3 | Nr1d2 | 474 | 0.534 | 0.0524 | Yes |
| 4 | Lamc1 | 512 | 0.525 | 0.0746 | Yes |
| 5 | Dab2 | 568 | 0.509 | 0.0951 | Yes |
| 6 | Dusp1 | 891 | 0.454 | 0.0995 | Yes |
| 7 | Pias3 | 905 | 0.452 | 0.1195 | Yes |
| 8 | Dyrk1a | 2150 | 0.319 | 0.0708 | No |
| 9 | Nek7 | 2192 | 0.317 | 0.0832 | No |
| 10 | Rnd3 | 2457 | 0.299 | 0.0834 | No |
| 11 | Tfpi | 2705 | 0.281 | 0.0837 | No |
| 12 | Fhl2 | 2726 | 0.280 | 0.0955 | No |
| 13 | Lpar1 | 2992 | 0.263 | 0.0941 | No |
| 14 | Pparg | 3188 | 0.251 | 0.0956 | No |
| 15 | Kalrn | 3729 | 0.221 | 0.0783 | No |
| 16 | Cap2 | 3846 | 0.215 | 0.0822 | No |
| 17 | Notch2 | 3918 | 0.211 | 0.0883 | No |
| 18 | Mta1 | 3946 | 0.210 | 0.0965 | No |
| 19 | Irs1 | 4340 | 0.191 | 0.0853 | No |
| 20 | Bmpr1a | 4594 | 0.179 | 0.0806 | No |
| 21 | Sipa1l1 | 4603 | 0.178 | 0.0883 | No |
| 22 | Gja1 | 4785 | 0.171 | 0.0869 | No |
| 23 | Synj2 | 5014 | 0.160 | 0.0827 | No |
| 24 | Ythdc1 | 5373 | 0.143 | 0.0710 | No |
| 25 | Smad3 | 5407 | 0.141 | 0.0758 | No |
| 26 | Rasa2 | 5438 | 0.139 | 0.0806 | No |
| 27 | Mios | 5488 | 0.137 | 0.0844 | No |
| 28 | Abcc1 | 5527 | 0.136 | 0.0887 | No |
| 29 | Pten | 5551 | 0.135 | 0.0937 | No |
| 30 | Akt3 | 5558 | 0.135 | 0.0996 | No |
| 31 | Cdkn1b | 5586 | 0.134 | 0.1043 | No |
| 32 | Igf1r | 5840 | 0.125 | 0.0972 | No |
| 33 | Col1a2 | 5858 | 0.124 | 0.1020 | No |
| 34 | Pik3cd | 5968 | 0.120 | 0.1019 | No |
| 35 | Mapk14 | 6239 | 0.112 | 0.0933 | No |
| 36 | Scaf8 | 6411 | 0.105 | 0.0894 | No |
| 37 | Slc7a1 | 6476 | 0.102 | 0.0908 | No |
| 38 | Rbpms | 6511 | 0.101 | 0.0937 | No |
| 39 | Magi2 | 6595 | 0.097 | 0.0939 | No |
| 40 | Atp2c1 | 6599 | 0.097 | 0.0982 | No |
| 41 | Phf3 | 6768 | 0.091 | 0.0938 | No |
| 42 | Insig1 | 6853 | 0.088 | 0.0936 | No |
| 43 | Anxa4 | 6935 | 0.085 | 0.0934 | No |
| 44 | Cdc42bpa | 6980 | 0.084 | 0.0950 | No |
| 45 | Cav1 | 7385 | 0.068 | 0.0775 | No |
| 46 | Syne1 | 7476 | 0.064 | 0.0759 | No |
| 47 | Bckdhb | 7515 | 0.063 | 0.0768 | No |
| 48 | Cdk13 | 7768 | 0.054 | 0.0665 | No |
| 49 | Aggf1 | 7791 | 0.053 | 0.0678 | No |
| 50 | Apbb2 | 7856 | 0.052 | 0.0669 | No |
| 51 | Spop | 7957 | 0.049 | 0.0640 | No |
| 52 | Mgmt | 8206 | 0.039 | 0.0532 | No |
| 53 | Itgb3 | 8227 | 0.038 | 0.0539 | No |
| 54 | Prkar2b | 8291 | 0.037 | 0.0524 | No |
| 55 | Atrx | 8347 | 0.035 | 0.0512 | No |
| 56 | Togaram1 | 8770 | 0.019 | 0.0306 | No |
| 57 | Acvr2a | 8922 | 0.013 | 0.0235 | No |
| 58 | Map2k5 | 8973 | 0.011 | 0.0215 | No |
| 59 | Ica1 | 8983 | 0.011 | 0.0215 | No |
| 60 | Amph | 8989 | 0.010 | 0.0217 | No |
| 61 | Tgfbr3 | 9180 | 0.004 | 0.0123 | No |
| 62 | Kcnma1 | 9201 | 0.003 | 0.0114 | No |
| 63 | Ldlr | 9260 | 0.001 | 0.0085 | No |
| 64 | Col1a1 | 9280 | 0.000 | 0.0075 | No |
| 65 | Schip1 | 9348 | -0.000 | 0.0041 | No |
| 66 | Wdr37 | 9598 | -0.008 | -0.0082 | No |
| 67 | Id1 | 9798 | -0.017 | -0.0175 | No |
| 68 | Ptprm | 9870 | -0.020 | -0.0202 | No |
| 69 | Kit | 9907 | -0.021 | -0.0211 | No |
| 70 | Nipbl | 10007 | -0.025 | -0.0250 | No |
| 71 | Tent4a | 10084 | -0.027 | -0.0277 | No |
| 72 | Pdgfrb | 10086 | -0.027 | -0.0265 | No |
| 73 | Add3 | 10237 | -0.033 | -0.0326 | No |
| 74 | Sdc2 | 10281 | -0.035 | -0.0332 | No |
| 75 | Prkce | 10401 | -0.039 | -0.0375 | No |
| 76 | Map1b | 10436 | -0.040 | -0.0374 | No |
| 77 | Atp2b1 | 10503 | -0.043 | -0.0388 | No |
| 78 | Dlc1 | 10788 | -0.052 | -0.0508 | No |
| 79 | Plcb4 | 10821 | -0.054 | -0.0500 | No |
| 80 | Adgrl2 | 10939 | -0.058 | -0.0533 | No |
| 81 | Sfmbt1 | 11082 | -0.060 | -0.0578 | No |
| 82 | Nr3c1 | 11161 | -0.063 | -0.0589 | No |
| 83 | Mt1 | 11217 | -0.065 | -0.0587 | No |
| 84 | Pex14 | 11257 | -0.067 | -0.0576 | No |
| 85 | Dlg1 | 11436 | -0.074 | -0.0633 | No |
| 86 | Grk5 | 11477 | -0.076 | -0.0618 | No |
| 87 | Vav2 | 11537 | -0.078 | -0.0612 | No |
| 88 | Met | 11828 | -0.090 | -0.0719 | No |
| 89 | Pdlim5 | 11860 | -0.091 | -0.0693 | No |
| 90 | Celf2 | 11953 | -0.096 | -0.0696 | No |
| 91 | Anxa2 | 12214 | -0.107 | -0.0779 | No |
| 92 | Zmiz1 | 12256 | -0.108 | -0.0750 | No |
| 93 | Ptpn21 | 12297 | -0.110 | -0.0720 | No |
| 94 | Prdm2 | 12342 | -0.112 | -0.0692 | No |
| 95 | Vldlr | 12345 | -0.112 | -0.0641 | No |
| 96 | Arhgef9 | 12475 | -0.117 | -0.0653 | No |
| 97 | Bhlhe40 | 12709 | -0.126 | -0.0715 | No |
| 98 | Atp2b4 | 12898 | -0.134 | -0.0749 | No |
| 99 | Mrps31 | 13230 | -0.146 | -0.0850 | No |
| 100 | Mgll | 13292 | -0.149 | -0.0813 | No |
| 101 | Dmac2l | 13525 | -0.158 | -0.0859 | No |
| 102 | Ltbp1 | 13605 | -0.162 | -0.0825 | No |
| 103 | Atrn | 13619 | -0.163 | -0.0757 | No |
| 104 | Atxn1 | 14244 | -0.193 | -0.0986 | No |
| 105 | Dbp | 14359 | -0.199 | -0.0953 | No |
| 106 | Tgfbr2 | 14404 | -0.201 | -0.0883 | No |
| 107 | Gcnt1 | 14458 | -0.205 | -0.0816 | No |
| 108 | Nfkb1 | 14543 | -0.209 | -0.0764 | No |
| 109 | Fyn | 14820 | -0.222 | -0.0803 | No |
| 110 | Myc | 14831 | -0.223 | -0.0706 | No |
| 111 | Runx1 | 15045 | -0.233 | -0.0708 | No |
| 112 | Plpp3 | 15471 | -0.253 | -0.0808 | No |
| 113 | Smad7 | 15758 | -0.268 | -0.0831 | No |
| 114 | Pik3r3 | 16044 | -0.288 | -0.0844 | No |
| 115 | Prkca | 16429 | -0.312 | -0.0897 | No |
| 116 | Mmp16 | 16709 | -0.330 | -0.0888 | No |
| 117 | Cdon | 17138 | -0.361 | -0.0940 | No |
| 118 | F3 | 17195 | -0.365 | -0.0802 | No |
| 119 | Nrp1 | 17831 | -0.420 | -0.0933 | No |
| 120 | Sri | 17838 | -0.420 | -0.0744 | No |
| 121 | Ddah1 | 17953 | -0.430 | -0.0605 | No |
| 122 | Inpp4b | 17959 | -0.431 | -0.0410 | No |
| 123 | Col5a2 | 17993 | -0.434 | -0.0228 | No |
| 124 | Cited2 | 18176 | -0.453 | -0.0113 | No |
| 125 | Adora2b | 18230 | -0.459 | 0.0070 | No |
| 126 | Tjp1 | 18613 | -0.508 | 0.0108 | No |
| 127 | Cacna1a | 18699 | -0.520 | 0.0302 | No |
| 128 | Slc22a18 | 18745 | -0.530 | 0.0522 | No |