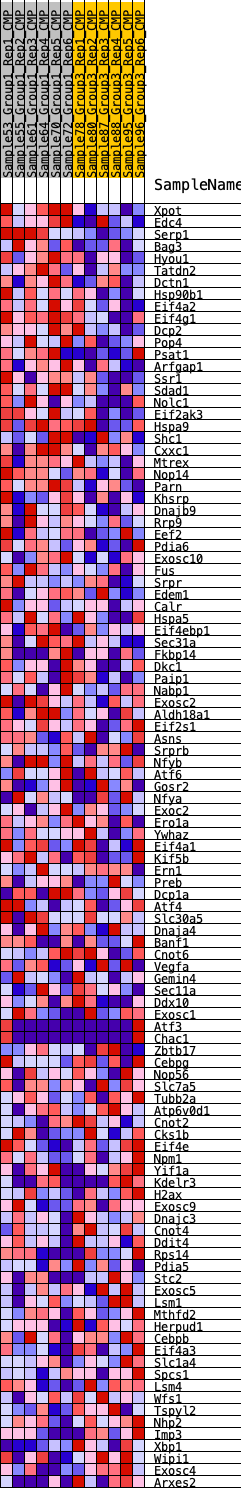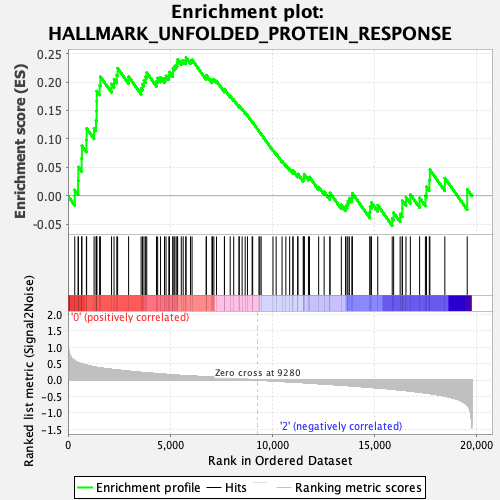
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group3.CMP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.24324133 |
| Normalized Enrichment Score (NES) | 0.979481 |
| Nominal p-value | 0.452 |
| FDR q-value | 0.90572125 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Xpot | 330 | 0.582 | 0.0104 | Yes |
| 2 | Edc4 | 496 | 0.529 | 0.0267 | Yes |
| 3 | Serp1 | 506 | 0.526 | 0.0509 | Yes |
| 4 | Bag3 | 668 | 0.492 | 0.0657 | Yes |
| 5 | Hyou1 | 680 | 0.490 | 0.0880 | Yes |
| 6 | Tatdn2 | 900 | 0.453 | 0.0981 | Yes |
| 7 | Dctn1 | 912 | 0.451 | 0.1186 | Yes |
| 8 | Hsp90b1 | 1277 | 0.400 | 0.1188 | Yes |
| 9 | Eif4a2 | 1371 | 0.388 | 0.1322 | Yes |
| 10 | Eif4g1 | 1399 | 0.384 | 0.1487 | Yes |
| 11 | Dcp2 | 1406 | 0.383 | 0.1663 | Yes |
| 12 | Pop4 | 1409 | 0.382 | 0.1841 | Yes |
| 13 | Psat1 | 1548 | 0.367 | 0.1942 | Yes |
| 14 | Arfgap1 | 1586 | 0.364 | 0.2093 | Yes |
| 15 | Ssr1 | 2132 | 0.321 | 0.1966 | Yes |
| 16 | Sdad1 | 2254 | 0.312 | 0.2050 | Yes |
| 17 | Nolc1 | 2390 | 0.303 | 0.2123 | Yes |
| 18 | Eif2ak3 | 2429 | 0.300 | 0.2244 | Yes |
| 19 | Hspa9 | 2967 | 0.265 | 0.2095 | Yes |
| 20 | Shc1 | 3580 | 0.229 | 0.1891 | Yes |
| 21 | Cxxc1 | 3650 | 0.225 | 0.1961 | Yes |
| 22 | Mtrex | 3721 | 0.222 | 0.2029 | Yes |
| 23 | Nop14 | 3800 | 0.217 | 0.2091 | Yes |
| 24 | Parn | 3856 | 0.214 | 0.2163 | Yes |
| 25 | Khsrp | 4333 | 0.192 | 0.2011 | Yes |
| 26 | Dnajb9 | 4389 | 0.189 | 0.2071 | Yes |
| 27 | Rrp9 | 4531 | 0.181 | 0.2084 | Yes |
| 28 | Eef2 | 4725 | 0.174 | 0.2067 | Yes |
| 29 | Pdia6 | 4800 | 0.170 | 0.2109 | Yes |
| 30 | Exosc10 | 4937 | 0.163 | 0.2116 | Yes |
| 31 | Fus | 4970 | 0.161 | 0.2176 | Yes |
| 32 | Srpr | 5134 | 0.154 | 0.2165 | Yes |
| 33 | Edem1 | 5136 | 0.154 | 0.2237 | Yes |
| 34 | Calr | 5205 | 0.151 | 0.2273 | Yes |
| 35 | Hspa5 | 5289 | 0.147 | 0.2299 | Yes |
| 36 | Eif4ebp1 | 5341 | 0.145 | 0.2341 | Yes |
| 37 | Sec31a | 5368 | 0.144 | 0.2395 | Yes |
| 38 | Fkbp14 | 5547 | 0.135 | 0.2367 | Yes |
| 39 | Dkc1 | 5640 | 0.132 | 0.2382 | Yes |
| 40 | Paip1 | 5769 | 0.127 | 0.2377 | Yes |
| 41 | Nabp1 | 5777 | 0.127 | 0.2432 | Yes |
| 42 | Exosc2 | 5997 | 0.119 | 0.2377 | No |
| 43 | Aldh18a1 | 6074 | 0.116 | 0.2392 | No |
| 44 | Eif2s1 | 6767 | 0.091 | 0.2083 | No |
| 45 | Asns | 6773 | 0.091 | 0.2123 | No |
| 46 | Srprb | 7048 | 0.081 | 0.2022 | No |
| 47 | Nfyb | 7075 | 0.080 | 0.2046 | No |
| 48 | Atf6 | 7142 | 0.078 | 0.2049 | No |
| 49 | Gosr2 | 7270 | 0.073 | 0.2018 | No |
| 50 | Nfya | 7658 | 0.058 | 0.1849 | No |
| 51 | Exoc2 | 7661 | 0.058 | 0.1875 | No |
| 52 | Ero1a | 7941 | 0.049 | 0.1756 | No |
| 53 | Ywhaz | 8113 | 0.043 | 0.1689 | No |
| 54 | Eif4a1 | 8373 | 0.033 | 0.1573 | No |
| 55 | Kif5b | 8381 | 0.033 | 0.1585 | No |
| 56 | Ern1 | 8525 | 0.028 | 0.1525 | No |
| 57 | Preb | 8674 | 0.023 | 0.1460 | No |
| 58 | Dcp1a | 8787 | 0.019 | 0.1412 | No |
| 59 | Atf4 | 9015 | 0.010 | 0.1301 | No |
| 60 | Slc30a5 | 9042 | 0.009 | 0.1292 | No |
| 61 | Dnaja4 | 9351 | -0.000 | 0.1136 | No |
| 62 | Banf1 | 9392 | -0.001 | 0.1116 | No |
| 63 | Cnot6 | 9457 | -0.003 | 0.1085 | No |
| 64 | Vegfa | 10036 | -0.025 | 0.0803 | No |
| 65 | Gemin4 | 10191 | -0.031 | 0.0739 | No |
| 66 | Sec11a | 10480 | -0.042 | 0.0612 | No |
| 67 | Ddx10 | 10667 | -0.048 | 0.0540 | No |
| 68 | Exosc1 | 10852 | -0.055 | 0.0472 | No |
| 69 | Atf3 | 11014 | -0.060 | 0.0418 | No |
| 70 | Chac1 | 11015 | -0.060 | 0.0446 | No |
| 71 | Zbtb17 | 11245 | -0.066 | 0.0361 | No |
| 72 | Cebpg | 11259 | -0.067 | 0.0386 | No |
| 73 | Nop56 | 11517 | -0.078 | 0.0291 | No |
| 74 | Slc7a5 | 11553 | -0.079 | 0.0310 | No |
| 75 | Tubb2a | 11554 | -0.079 | 0.0347 | No |
| 76 | Atp6v0d1 | 11563 | -0.079 | 0.0380 | No |
| 77 | Cnot2 | 11773 | -0.088 | 0.0315 | No |
| 78 | Cks1b | 11823 | -0.090 | 0.0332 | No |
| 79 | Eif4e | 12272 | -0.109 | 0.0155 | No |
| 80 | Npm1 | 12540 | -0.119 | 0.0075 | No |
| 81 | Yif1a | 12824 | -0.131 | -0.0008 | No |
| 82 | Kdelr3 | 12826 | -0.131 | 0.0053 | No |
| 83 | H2ax | 13383 | -0.153 | -0.0158 | No |
| 84 | Exosc9 | 13589 | -0.161 | -0.0187 | No |
| 85 | Dnajc3 | 13662 | -0.165 | -0.0147 | No |
| 86 | Cnot4 | 13706 | -0.167 | -0.0091 | No |
| 87 | Ddit4 | 13776 | -0.170 | -0.0046 | No |
| 88 | Rps14 | 13899 | -0.175 | -0.0026 | No |
| 89 | Pdia5 | 13925 | -0.177 | 0.0044 | No |
| 90 | Stc2 | 14778 | -0.220 | -0.0287 | No |
| 91 | Exosc5 | 14791 | -0.221 | -0.0190 | No |
| 92 | Lsm1 | 14858 | -0.224 | -0.0118 | No |
| 93 | Mthfd2 | 15165 | -0.238 | -0.0163 | No |
| 94 | Herpud1 | 15874 | -0.277 | -0.0393 | No |
| 95 | Cebpb | 15943 | -0.281 | -0.0296 | No |
| 96 | Eif4a3 | 16262 | -0.301 | -0.0317 | No |
| 97 | Slc1a4 | 16366 | -0.308 | -0.0226 | No |
| 98 | Spcs1 | 16368 | -0.308 | -0.0083 | No |
| 99 | Lsm4 | 16547 | -0.319 | -0.0024 | No |
| 100 | Wfs1 | 16762 | -0.334 | 0.0023 | No |
| 101 | Tspyl2 | 17215 | -0.366 | -0.0035 | No |
| 102 | Nhp2 | 17498 | -0.390 | 0.0003 | No |
| 103 | Imp3 | 17552 | -0.395 | 0.0161 | No |
| 104 | Xbp1 | 17691 | -0.407 | 0.0281 | No |
| 105 | Wipi1 | 17718 | -0.410 | 0.0459 | No |
| 106 | Exosc4 | 18450 | -0.482 | 0.0313 | No |
| 107 | Arxes2 | 19546 | -0.766 | 0.0114 | No |