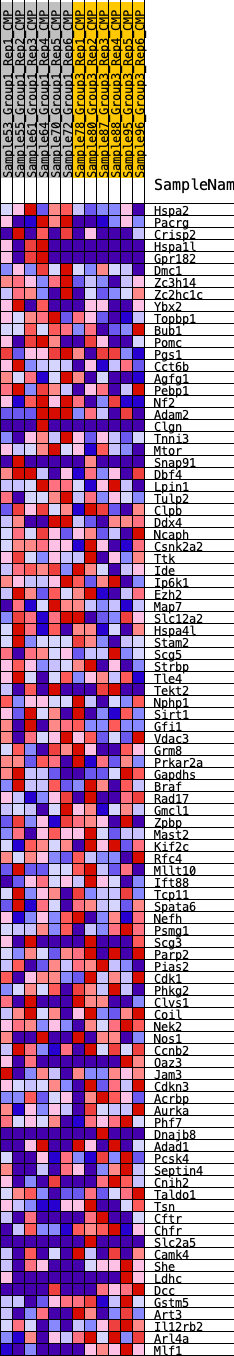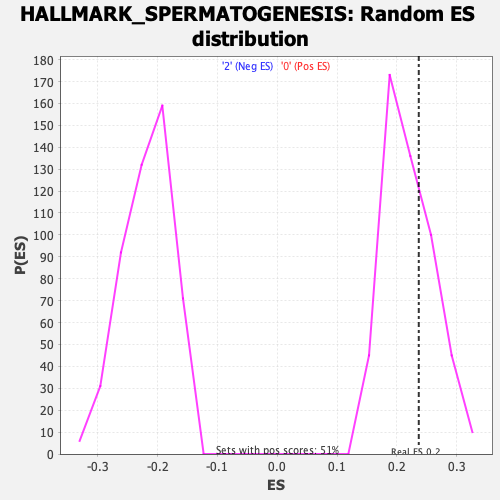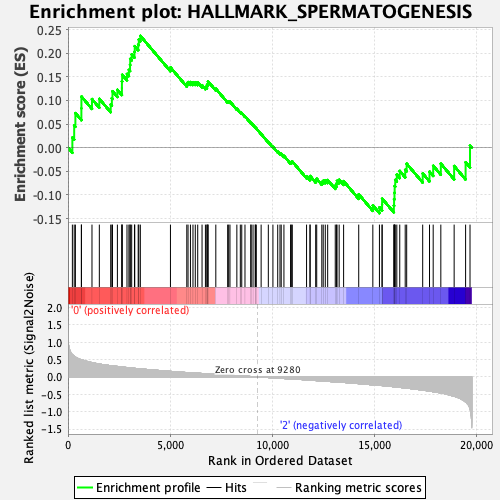
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group3.CMP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.23651621 |
| Normalized Enrichment Score (NES) | 1.0773085 |
| Nominal p-value | 0.33988214 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.973 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hspa2 | 214 | 0.639 | 0.0215 | Yes |
| 2 | Pacrg | 303 | 0.594 | 0.0471 | Yes |
| 3 | Crisp2 | 362 | 0.571 | 0.0730 | Yes |
| 4 | Hspa1l | 655 | 0.493 | 0.0832 | Yes |
| 5 | Gpr182 | 658 | 0.493 | 0.1081 | Yes |
| 6 | Dmc1 | 1174 | 0.411 | 0.1028 | Yes |
| 7 | Zc3h14 | 1534 | 0.369 | 0.1032 | Yes |
| 8 | Zc2hc1c | 2091 | 0.324 | 0.0914 | Yes |
| 9 | Ybx2 | 2148 | 0.319 | 0.1047 | Yes |
| 10 | Topbp1 | 2178 | 0.318 | 0.1193 | Yes |
| 11 | Bub1 | 2418 | 0.302 | 0.1224 | Yes |
| 12 | Pomc | 2639 | 0.287 | 0.1258 | Yes |
| 13 | Pgs1 | 2640 | 0.287 | 0.1404 | Yes |
| 14 | Cct6b | 2653 | 0.286 | 0.1543 | Yes |
| 15 | Agfg1 | 2888 | 0.270 | 0.1560 | Yes |
| 16 | Pebp1 | 2982 | 0.264 | 0.1647 | Yes |
| 17 | Nf2 | 3038 | 0.261 | 0.1751 | Yes |
| 18 | Adam2 | 3046 | 0.260 | 0.1879 | Yes |
| 19 | Clgn | 3118 | 0.255 | 0.1973 | Yes |
| 20 | Tnni3 | 3255 | 0.247 | 0.2029 | Yes |
| 21 | Mtor | 3268 | 0.246 | 0.2147 | Yes |
| 22 | Snap91 | 3437 | 0.237 | 0.2182 | Yes |
| 23 | Dbf4 | 3467 | 0.235 | 0.2286 | Yes |
| 24 | Lpin1 | 3542 | 0.231 | 0.2365 | Yes |
| 25 | Tulp2 | 5016 | 0.160 | 0.1697 | No |
| 26 | Clpb | 5816 | 0.126 | 0.1355 | No |
| 27 | Ddx4 | 5877 | 0.123 | 0.1387 | No |
| 28 | Ncaph | 5995 | 0.119 | 0.1388 | No |
| 29 | Csnk2a2 | 6119 | 0.114 | 0.1383 | No |
| 30 | Ttk | 6232 | 0.112 | 0.1383 | No |
| 31 | Ide | 6351 | 0.108 | 0.1378 | No |
| 32 | Ip6k1 | 6563 | 0.099 | 0.1321 | No |
| 33 | Ezh2 | 6728 | 0.093 | 0.1284 | No |
| 34 | Map7 | 6801 | 0.090 | 0.1293 | No |
| 35 | Slc12a2 | 6812 | 0.090 | 0.1334 | No |
| 36 | Hspa4l | 6851 | 0.088 | 0.1359 | No |
| 37 | Stam2 | 6855 | 0.088 | 0.1402 | No |
| 38 | Scg5 | 7237 | 0.074 | 0.1246 | No |
| 39 | Strbp | 7817 | 0.053 | 0.0979 | No |
| 40 | Tle4 | 7867 | 0.051 | 0.0980 | No |
| 41 | Tekt2 | 7930 | 0.049 | 0.0973 | No |
| 42 | Nphp1 | 8267 | 0.038 | 0.0822 | No |
| 43 | Sirt1 | 8437 | 0.031 | 0.0751 | No |
| 44 | Gfi1 | 8507 | 0.029 | 0.0731 | No |
| 45 | Vdac3 | 8665 | 0.023 | 0.0663 | No |
| 46 | Grm8 | 8948 | 0.012 | 0.0525 | No |
| 47 | Prkar2a | 8968 | 0.011 | 0.0521 | No |
| 48 | Gapdhs | 9059 | 0.008 | 0.0480 | No |
| 49 | Braf | 9165 | 0.005 | 0.0429 | No |
| 50 | Rad17 | 9178 | 0.004 | 0.0425 | No |
| 51 | Gmcl1 | 9220 | 0.003 | 0.0406 | No |
| 52 | Zpbp | 9456 | -0.003 | 0.0288 | No |
| 53 | Mast2 | 9808 | -0.017 | 0.0118 | No |
| 54 | Kif2c | 10033 | -0.025 | 0.0017 | No |
| 55 | Rfc4 | 10268 | -0.034 | -0.0084 | No |
| 56 | Mllt10 | 10379 | -0.038 | -0.0121 | No |
| 57 | Ift88 | 10439 | -0.040 | -0.0130 | No |
| 58 | Tcp11 | 10572 | -0.045 | -0.0175 | No |
| 59 | Spata6 | 10897 | -0.056 | -0.0311 | No |
| 60 | Nefh | 10935 | -0.058 | -0.0300 | No |
| 61 | Psmg1 | 10982 | -0.059 | -0.0294 | No |
| 62 | Scg3 | 11680 | -0.084 | -0.0606 | No |
| 63 | Parp2 | 11849 | -0.091 | -0.0645 | No |
| 64 | Pias2 | 11853 | -0.091 | -0.0600 | No |
| 65 | Cdk1 | 12127 | -0.103 | -0.0687 | No |
| 66 | Phkg2 | 12177 | -0.105 | -0.0658 | No |
| 67 | Clvs1 | 12427 | -0.116 | -0.0726 | No |
| 68 | Coil | 12495 | -0.118 | -0.0701 | No |
| 69 | Nek2 | 12597 | -0.122 | -0.0690 | No |
| 70 | Nos1 | 12714 | -0.126 | -0.0685 | No |
| 71 | Ccnb2 | 13085 | -0.139 | -0.0803 | No |
| 72 | Oaz3 | 13143 | -0.142 | -0.0760 | No |
| 73 | Jam3 | 13172 | -0.144 | -0.0701 | No |
| 74 | Cdkn3 | 13276 | -0.148 | -0.0678 | No |
| 75 | Acrbp | 13494 | -0.157 | -0.0709 | No |
| 76 | Aurka | 14234 | -0.193 | -0.0987 | No |
| 77 | Phf7 | 14925 | -0.228 | -0.1222 | No |
| 78 | Dnajb8 | 15248 | -0.242 | -0.1263 | No |
| 79 | Adad1 | 15378 | -0.248 | -0.1203 | No |
| 80 | Pcsk4 | 15380 | -0.248 | -0.1078 | No |
| 81 | Septin4 | 15954 | -0.282 | -0.1227 | No |
| 82 | Cnih2 | 15966 | -0.284 | -0.1089 | No |
| 83 | Taldo1 | 15985 | -0.285 | -0.0953 | No |
| 84 | Tsn | 15994 | -0.285 | -0.0813 | No |
| 85 | Cftr | 16022 | -0.287 | -0.0681 | No |
| 86 | Chfr | 16099 | -0.291 | -0.0572 | No |
| 87 | Slc2a5 | 16243 | -0.300 | -0.0493 | No |
| 88 | Camk4 | 16510 | -0.317 | -0.0468 | No |
| 89 | She | 16582 | -0.322 | -0.0340 | No |
| 90 | Ldhc | 17366 | -0.380 | -0.0546 | No |
| 91 | Dcc | 17700 | -0.408 | -0.0508 | No |
| 92 | Gstm5 | 17880 | -0.426 | -0.0384 | No |
| 93 | Art3 | 18254 | -0.461 | -0.0340 | No |
| 94 | Il12rb2 | 18906 | -0.556 | -0.0389 | No |
| 95 | Arl4a | 19467 | -0.717 | -0.0310 | No |
| 96 | Mlf1 | 19683 | -0.916 | 0.0045 | No |