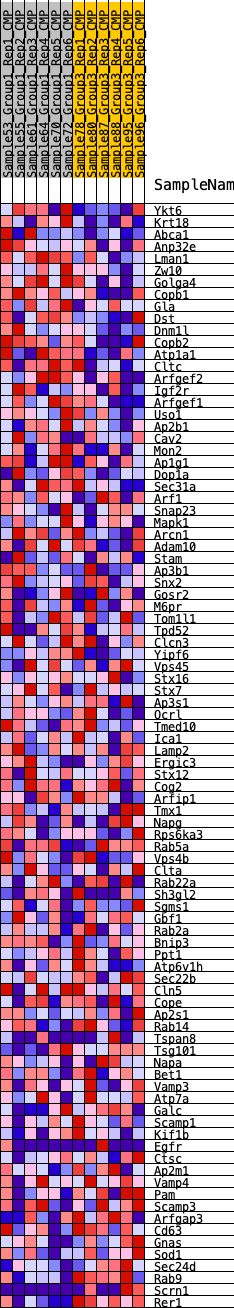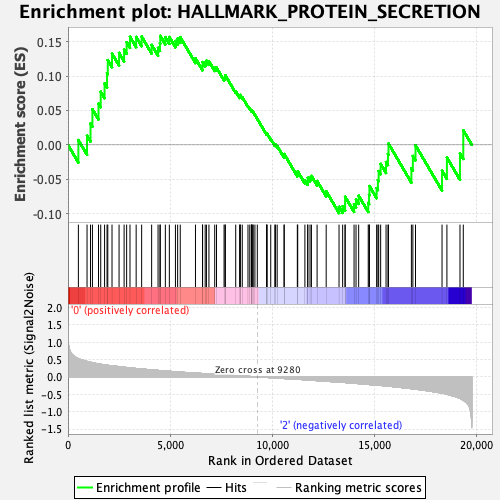
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group3.CMP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.1584581 |
| Normalized Enrichment Score (NES) | 0.7271463 |
| Nominal p-value | 0.7656904 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ykt6 | 508 | 0.526 | 0.0070 | Yes |
| 2 | Krt18 | 932 | 0.447 | 0.0134 | Yes |
| 3 | Abca1 | 1102 | 0.422 | 0.0311 | Yes |
| 4 | Anp32e | 1196 | 0.408 | 0.0519 | Yes |
| 5 | Lman1 | 1494 | 0.373 | 0.0600 | Yes |
| 6 | Zw10 | 1602 | 0.363 | 0.0772 | Yes |
| 7 | Golga4 | 1791 | 0.347 | 0.0893 | Yes |
| 8 | Copb1 | 1912 | 0.337 | 0.1042 | Yes |
| 9 | Gla | 1948 | 0.334 | 0.1233 | Yes |
| 10 | Dst | 2155 | 0.318 | 0.1327 | Yes |
| 11 | Dnm1l | 2500 | 0.296 | 0.1337 | Yes |
| 12 | Copb2 | 2747 | 0.278 | 0.1386 | Yes |
| 13 | Atp1a1 | 2871 | 0.271 | 0.1492 | Yes |
| 14 | Cltc | 3035 | 0.261 | 0.1572 | Yes |
| 15 | Arfgef2 | 3337 | 0.241 | 0.1569 | Yes |
| 16 | Igf2r | 3606 | 0.227 | 0.1575 | Yes |
| 17 | Arfgef1 | 4095 | 0.203 | 0.1454 | Yes |
| 18 | Uso1 | 4409 | 0.187 | 0.1411 | Yes |
| 19 | Ap2b1 | 4503 | 0.183 | 0.1478 | Yes |
| 20 | Cav2 | 4518 | 0.182 | 0.1585 | Yes |
| 21 | Mon2 | 4766 | 0.172 | 0.1566 | No |
| 22 | Ap1g1 | 4962 | 0.162 | 0.1568 | No |
| 23 | Dop1a | 5260 | 0.149 | 0.1510 | No |
| 24 | Sec31a | 5368 | 0.144 | 0.1545 | No |
| 25 | Arf1 | 5496 | 0.137 | 0.1566 | No |
| 26 | Snap23 | 6241 | 0.112 | 0.1258 | No |
| 27 | Mapk1 | 6586 | 0.098 | 0.1144 | No |
| 28 | Arcn1 | 6590 | 0.098 | 0.1204 | No |
| 29 | Adam10 | 6716 | 0.093 | 0.1198 | No |
| 30 | Stam | 6778 | 0.091 | 0.1224 | No |
| 31 | Ap3b1 | 6900 | 0.086 | 0.1216 | No |
| 32 | Snx2 | 7183 | 0.077 | 0.1121 | No |
| 33 | Gosr2 | 7270 | 0.073 | 0.1123 | No |
| 34 | M6pr | 7641 | 0.058 | 0.0971 | No |
| 35 | Tom1l1 | 7692 | 0.057 | 0.0981 | No |
| 36 | Tpd52 | 7703 | 0.056 | 0.1012 | No |
| 37 | Clcn3 | 8211 | 0.039 | 0.0778 | No |
| 38 | Yipf6 | 8405 | 0.032 | 0.0700 | No |
| 39 | Vps45 | 8430 | 0.031 | 0.0708 | No |
| 40 | Stx16 | 8432 | 0.031 | 0.0726 | No |
| 41 | Stx7 | 8532 | 0.028 | 0.0694 | No |
| 42 | Ap3s1 | 8812 | 0.017 | 0.0563 | No |
| 43 | Ocrl | 8900 | 0.014 | 0.0527 | No |
| 44 | Tmed10 | 8978 | 0.011 | 0.0495 | No |
| 45 | Ica1 | 8983 | 0.011 | 0.0500 | No |
| 46 | Lamp2 | 9000 | 0.010 | 0.0498 | No |
| 47 | Ergic3 | 9043 | 0.009 | 0.0482 | No |
| 48 | Stx12 | 9066 | 0.008 | 0.0476 | No |
| 49 | Cog2 | 9151 | 0.005 | 0.0437 | No |
| 50 | Arfip1 | 9269 | 0.000 | 0.0377 | No |
| 51 | Tmx1 | 9725 | -0.014 | 0.0155 | No |
| 52 | Napg | 9730 | -0.014 | 0.0162 | No |
| 53 | Rps6ka3 | 9746 | -0.015 | 0.0163 | No |
| 54 | Rab5a | 9926 | -0.021 | 0.0086 | No |
| 55 | Vps4b | 10126 | -0.029 | 0.0002 | No |
| 56 | Clta | 10163 | -0.030 | 0.0003 | No |
| 57 | Rab22a | 10242 | -0.033 | -0.0016 | No |
| 58 | Sh3gl2 | 10580 | -0.045 | -0.0159 | No |
| 59 | Sgms1 | 10585 | -0.046 | -0.0133 | No |
| 60 | Gbf1 | 11228 | -0.066 | -0.0418 | No |
| 61 | Rab2a | 11247 | -0.067 | -0.0385 | No |
| 62 | Bnip3 | 11599 | -0.080 | -0.0514 | No |
| 63 | Ppt1 | 11734 | -0.086 | -0.0528 | No |
| 64 | Atp6v1h | 11743 | -0.086 | -0.0478 | No |
| 65 | Sec22b | 11850 | -0.091 | -0.0475 | No |
| 66 | Cln5 | 11919 | -0.094 | -0.0451 | No |
| 67 | Cope | 12196 | -0.106 | -0.0525 | No |
| 68 | Ap2s1 | 12641 | -0.124 | -0.0674 | No |
| 69 | Rab14 | 13270 | -0.148 | -0.0901 | No |
| 70 | Tspan8 | 13447 | -0.156 | -0.0893 | No |
| 71 | Tsg101 | 13567 | -0.160 | -0.0853 | No |
| 72 | Napa | 13568 | -0.160 | -0.0753 | No |
| 73 | Bet1 | 14006 | -0.181 | -0.0862 | No |
| 74 | Vamp3 | 14102 | -0.186 | -0.0794 | No |
| 75 | Atp7a | 14231 | -0.192 | -0.0739 | No |
| 76 | Galc | 14700 | -0.216 | -0.0842 | No |
| 77 | Scamp1 | 14742 | -0.218 | -0.0727 | No |
| 78 | Kif1b | 14759 | -0.219 | -0.0598 | No |
| 79 | Egfr | 15111 | -0.237 | -0.0629 | No |
| 80 | Ctsc | 15174 | -0.239 | -0.0512 | No |
| 81 | Ap2m1 | 15212 | -0.240 | -0.0380 | No |
| 82 | Vamp4 | 15308 | -0.244 | -0.0277 | No |
| 83 | Pam | 15572 | -0.258 | -0.0249 | No |
| 84 | Scamp3 | 15665 | -0.262 | -0.0132 | No |
| 85 | Arfgap3 | 15692 | -0.264 | 0.0019 | No |
| 86 | Cd63 | 16813 | -0.338 | -0.0339 | No |
| 87 | Gnas | 16885 | -0.343 | -0.0161 | No |
| 88 | Sod1 | 17014 | -0.352 | -0.0006 | No |
| 89 | Sec24d | 18313 | -0.467 | -0.0374 | No |
| 90 | Rab9 | 18552 | -0.500 | -0.0183 | No |
| 91 | Scrn1 | 19192 | -0.615 | -0.0124 | No |
| 92 | Rer1 | 19354 | -0.670 | 0.0212 | No |