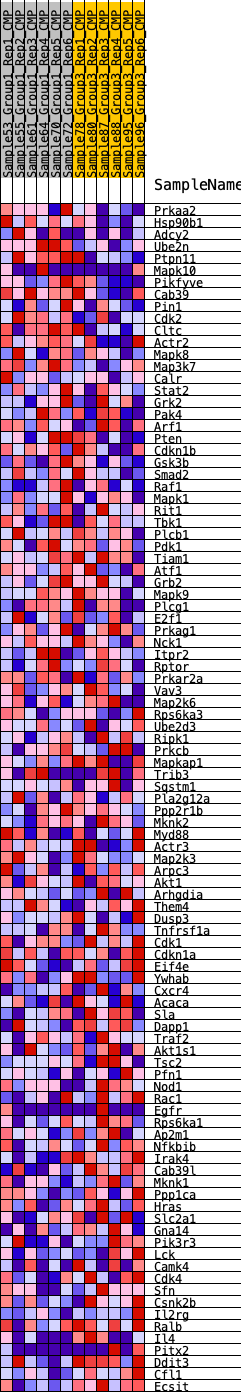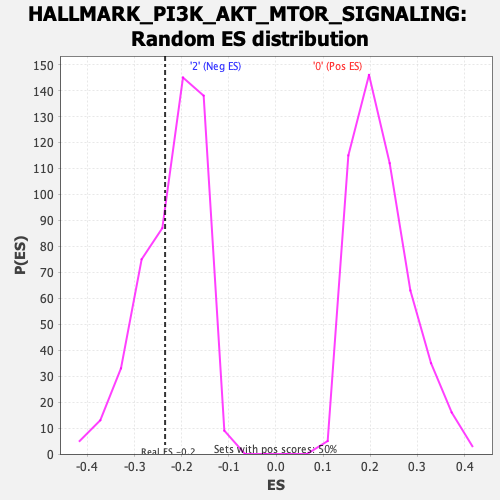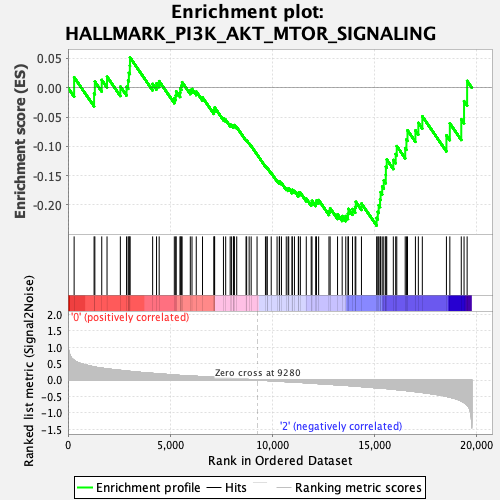
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group3.CMP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.235218 |
| Normalized Enrichment Score (NES) | -1.0653104 |
| Nominal p-value | 0.36237624 |
| FDR q-value | 0.845023 |
| FWER p-Value | 0.991 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Prkaa2 | 300 | 0.596 | 0.0178 | No |
| 2 | Hsp90b1 | 1277 | 0.400 | -0.0096 | No |
| 3 | Adcy2 | 1315 | 0.396 | 0.0105 | No |
| 4 | Ube2n | 1652 | 0.359 | 0.0133 | No |
| 5 | Ptpn11 | 1911 | 0.337 | 0.0189 | No |
| 6 | Mapk10 | 2563 | 0.291 | 0.0020 | No |
| 7 | Pikfyve | 2866 | 0.271 | 0.0017 | No |
| 8 | Cab39 | 2943 | 0.267 | 0.0126 | No |
| 9 | Pin1 | 2981 | 0.264 | 0.0254 | No |
| 10 | Cdk2 | 3028 | 0.261 | 0.0375 | No |
| 11 | Cltc | 3035 | 0.261 | 0.0517 | No |
| 12 | Actr2 | 4144 | 0.200 | 0.0065 | No |
| 13 | Mapk8 | 4335 | 0.191 | 0.0075 | No |
| 14 | Map3k7 | 4464 | 0.184 | 0.0112 | No |
| 15 | Calr | 5205 | 0.151 | -0.0180 | No |
| 16 | Stat2 | 5273 | 0.148 | -0.0132 | No |
| 17 | Grk2 | 5296 | 0.147 | -0.0062 | No |
| 18 | Pak4 | 5489 | 0.137 | -0.0083 | No |
| 19 | Arf1 | 5496 | 0.137 | -0.0010 | No |
| 20 | Pten | 5551 | 0.135 | 0.0037 | No |
| 21 | Cdkn1b | 5586 | 0.134 | 0.0094 | No |
| 22 | Gsk3b | 5988 | 0.119 | -0.0044 | No |
| 23 | Smad2 | 6071 | 0.116 | -0.0021 | No |
| 24 | Raf1 | 6280 | 0.111 | -0.0065 | No |
| 25 | Mapk1 | 6586 | 0.098 | -0.0166 | No |
| 26 | Rit1 | 7140 | 0.078 | -0.0404 | No |
| 27 | Tbk1 | 7164 | 0.078 | -0.0372 | No |
| 28 | Plcb1 | 7180 | 0.077 | -0.0337 | No |
| 29 | Pdk1 | 7611 | 0.059 | -0.0523 | No |
| 30 | Tiam1 | 7720 | 0.056 | -0.0547 | No |
| 31 | Atf1 | 7940 | 0.049 | -0.0631 | No |
| 32 | Grb2 | 8000 | 0.047 | -0.0635 | No |
| 33 | Mapk9 | 8097 | 0.043 | -0.0660 | No |
| 34 | Plcg1 | 8111 | 0.043 | -0.0642 | No |
| 35 | E2f1 | 8147 | 0.042 | -0.0637 | No |
| 36 | Prkag1 | 8261 | 0.038 | -0.0674 | No |
| 37 | Nck1 | 8717 | 0.021 | -0.0893 | No |
| 38 | Itpr2 | 8747 | 0.020 | -0.0897 | No |
| 39 | Rptor | 8869 | 0.015 | -0.0950 | No |
| 40 | Prkar2a | 8968 | 0.011 | -0.0993 | No |
| 41 | Vav3 | 9256 | 0.001 | -0.1139 | No |
| 42 | Map2k6 | 9667 | -0.011 | -0.1341 | No |
| 43 | Rps6ka3 | 9746 | -0.015 | -0.1372 | No |
| 44 | Ube2d3 | 9747 | -0.015 | -0.1364 | No |
| 45 | Ripk1 | 9950 | -0.022 | -0.1454 | No |
| 46 | Prkcb | 10236 | -0.033 | -0.1581 | No |
| 47 | Mapkap1 | 10344 | -0.037 | -0.1615 | No |
| 48 | Trib3 | 10358 | -0.038 | -0.1600 | No |
| 49 | Sqstm1 | 10451 | -0.041 | -0.1624 | No |
| 50 | Pla2g12a | 10686 | -0.049 | -0.1716 | No |
| 51 | Ppp2r1b | 10774 | -0.052 | -0.1732 | No |
| 52 | Mknk2 | 10807 | -0.053 | -0.1719 | No |
| 53 | Myd88 | 10969 | -0.058 | -0.1768 | No |
| 54 | Actr3 | 10980 | -0.059 | -0.1740 | No |
| 55 | Map2k3 | 11087 | -0.060 | -0.1761 | No |
| 56 | Arpc3 | 11277 | -0.068 | -0.1819 | No |
| 57 | Akt1 | 11292 | -0.068 | -0.1789 | No |
| 58 | Arhgdia | 11377 | -0.072 | -0.1791 | No |
| 59 | Them4 | 11666 | -0.083 | -0.1892 | No |
| 60 | Dusp3 | 11907 | -0.094 | -0.1962 | No |
| 61 | Tnfrsf1a | 11946 | -0.096 | -0.1928 | No |
| 62 | Cdk1 | 12127 | -0.103 | -0.1962 | No |
| 63 | Cdkn1a | 12166 | -0.105 | -0.1923 | No |
| 64 | Eif4e | 12272 | -0.109 | -0.1916 | No |
| 65 | Ywhab | 12777 | -0.129 | -0.2101 | No |
| 66 | Cxcr4 | 12834 | -0.131 | -0.2056 | No |
| 67 | Acaca | 13197 | -0.145 | -0.2160 | No |
| 68 | Sla | 13424 | -0.155 | -0.2189 | No |
| 69 | Dapp1 | 13600 | -0.162 | -0.2188 | No |
| 70 | Traf2 | 13708 | -0.167 | -0.2150 | No |
| 71 | Akt1s1 | 13729 | -0.168 | -0.2067 | No |
| 72 | Tsc2 | 13930 | -0.177 | -0.2070 | No |
| 73 | Pfn1 | 14059 | -0.184 | -0.2033 | No |
| 74 | Nod1 | 14092 | -0.186 | -0.1946 | No |
| 75 | Rac1 | 14368 | -0.200 | -0.1975 | No |
| 76 | Egfr | 15111 | -0.237 | -0.2221 | Yes |
| 77 | Rps6ka1 | 15168 | -0.238 | -0.2117 | Yes |
| 78 | Ap2m1 | 15212 | -0.240 | -0.2006 | Yes |
| 79 | Nfkbib | 15278 | -0.242 | -0.1904 | Yes |
| 80 | Irak4 | 15306 | -0.244 | -0.1783 | Yes |
| 81 | Cab39l | 15384 | -0.248 | -0.1684 | Yes |
| 82 | Mknk1 | 15461 | -0.252 | -0.1583 | Yes |
| 83 | Ppp1ca | 15559 | -0.258 | -0.1489 | Yes |
| 84 | Hras | 15564 | -0.258 | -0.1348 | Yes |
| 85 | Slc2a1 | 15606 | -0.259 | -0.1225 | Yes |
| 86 | Gna14 | 15928 | -0.281 | -0.1232 | Yes |
| 87 | Pik3r3 | 16044 | -0.288 | -0.1131 | Yes |
| 88 | Lck | 16097 | -0.291 | -0.0996 | Yes |
| 89 | Camk4 | 16510 | -0.317 | -0.1029 | Yes |
| 90 | Cdk4 | 16570 | -0.321 | -0.0881 | Yes |
| 91 | Sfn | 16622 | -0.325 | -0.0727 | Yes |
| 92 | Csnk2b | 17010 | -0.352 | -0.0728 | Yes |
| 93 | Il2rg | 17153 | -0.362 | -0.0599 | Yes |
| 94 | Ralb | 17348 | -0.379 | -0.0488 | Yes |
| 95 | Il4 | 18528 | -0.496 | -0.0812 | Yes |
| 96 | Pitx2 | 18695 | -0.519 | -0.0608 | Yes |
| 97 | Ddit3 | 19255 | -0.636 | -0.0539 | Yes |
| 98 | Cfl1 | 19391 | -0.682 | -0.0230 | Yes |
| 99 | Ecsit | 19542 | -0.761 | 0.0116 | Yes |